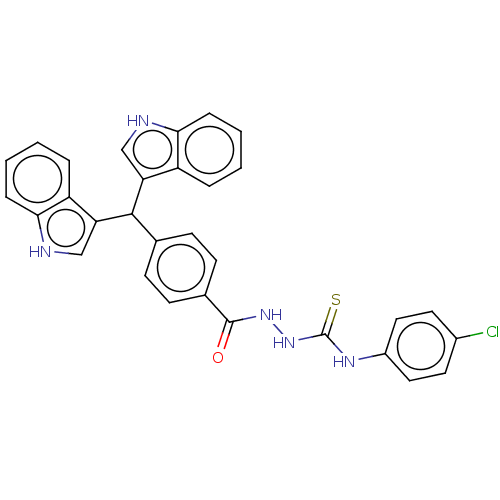
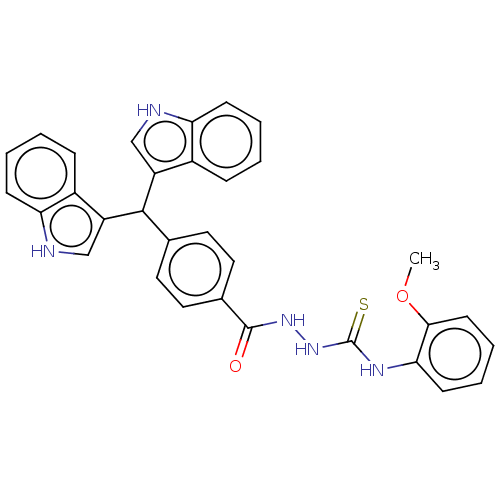
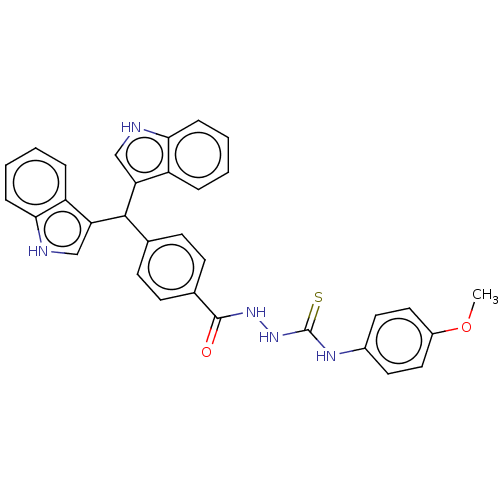
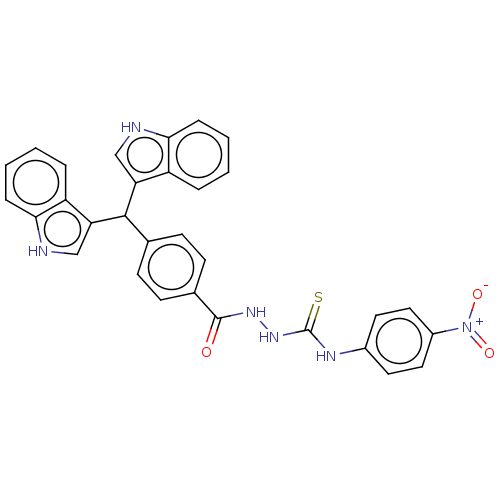
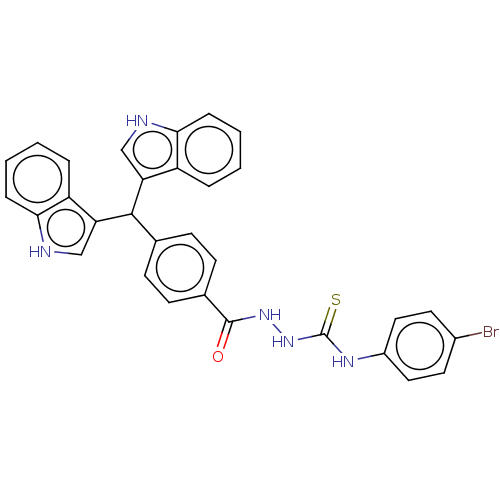
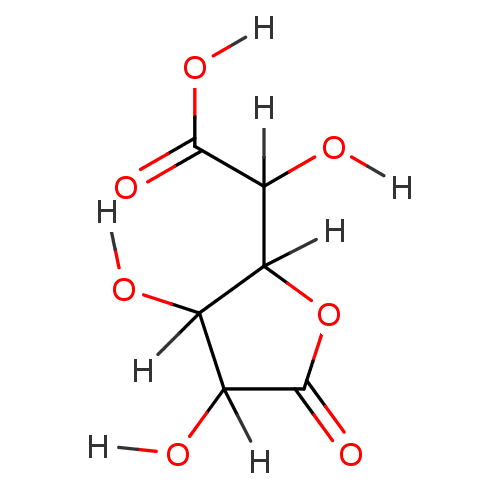
Report error Found 19 Enz. Inhib. hit(s) with all data for entry = 7415
Affinity DataIC50: 100nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 120nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 500nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 2.30E+3nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 2.90E+3nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 3.10E+3nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 5.70E+3nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 7.12E+3nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 8.50E+3nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 9.40E+3nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 1.16E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 1.47E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 2.14E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 2.81E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 3.32E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair
Affinity DataIC50: 4.84E+4nMAssay Description:β-Glucuronidase activity was determined by measuring the absorbance at 405 nm of p-nitrophenol formed from the substrate by the spectrophotometr...More data for this Ligand-Target Pair