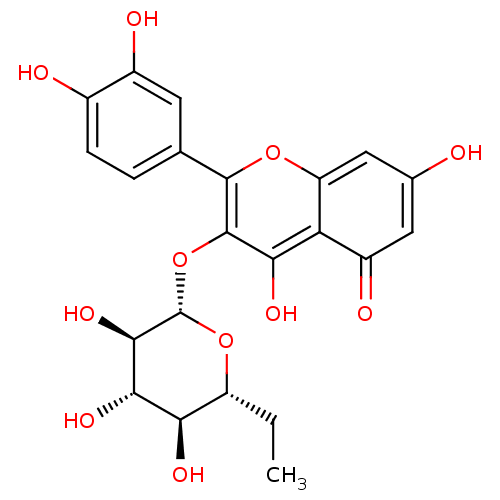
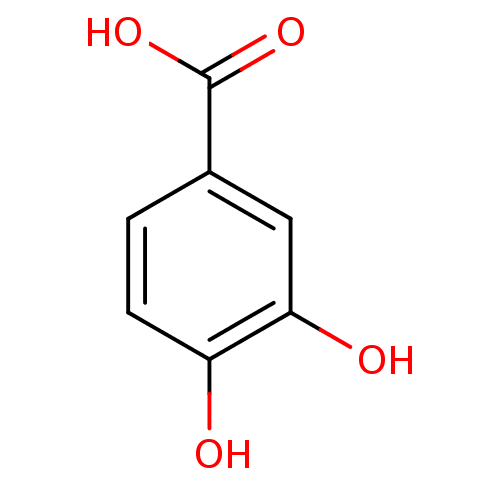
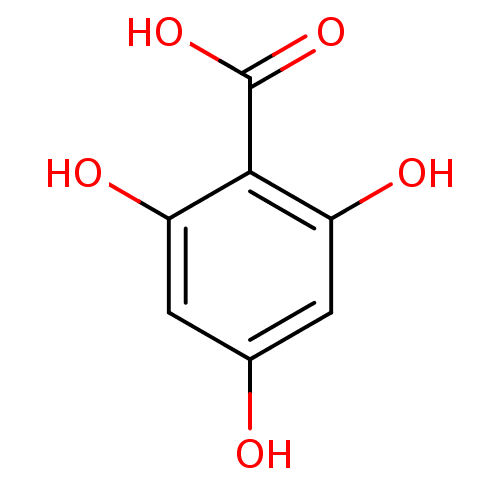
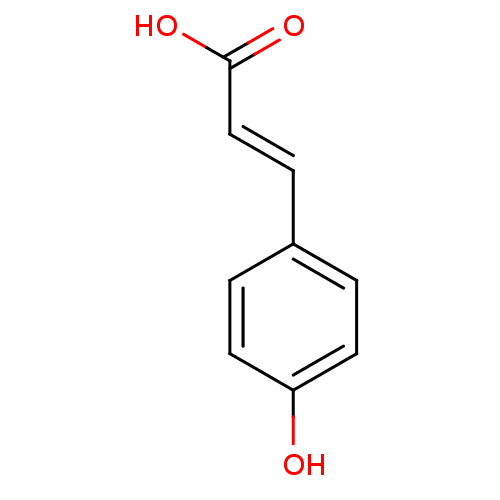
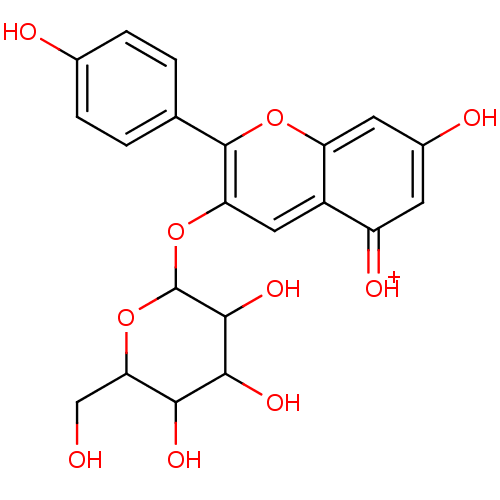
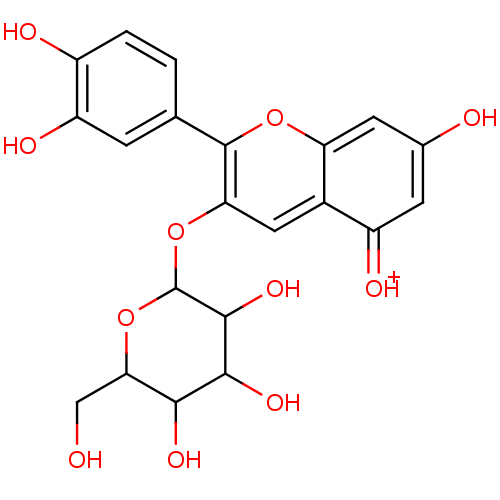
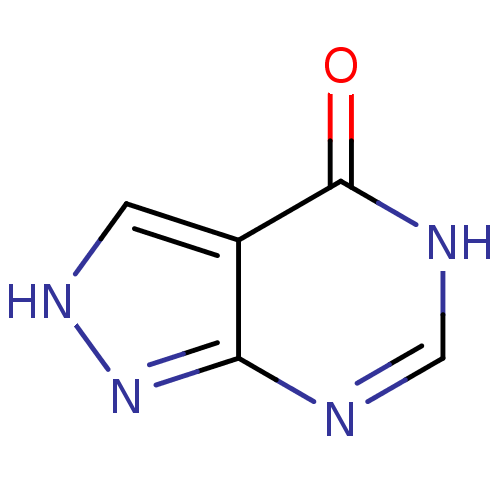
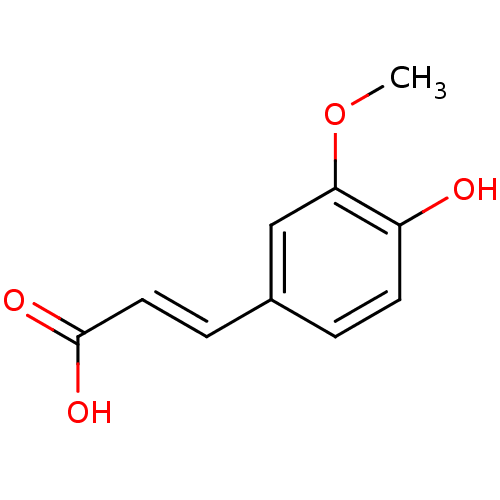
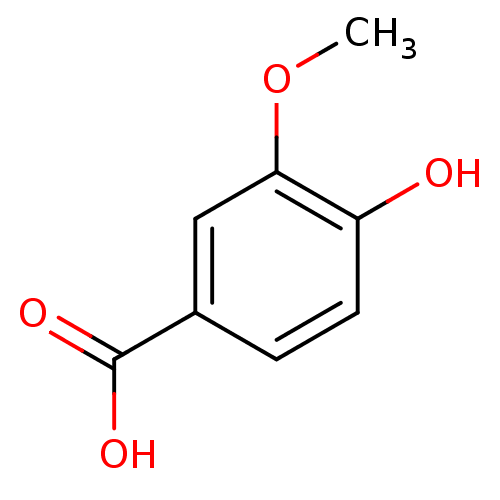
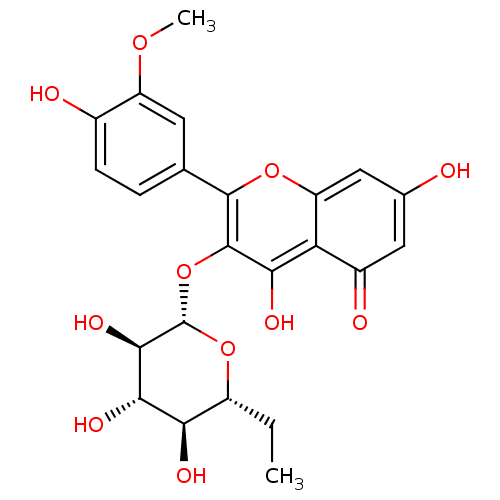
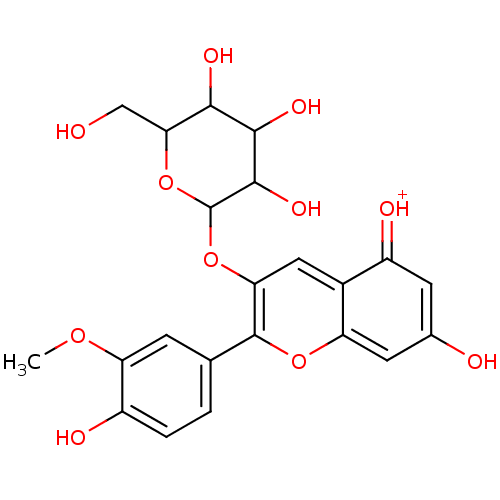
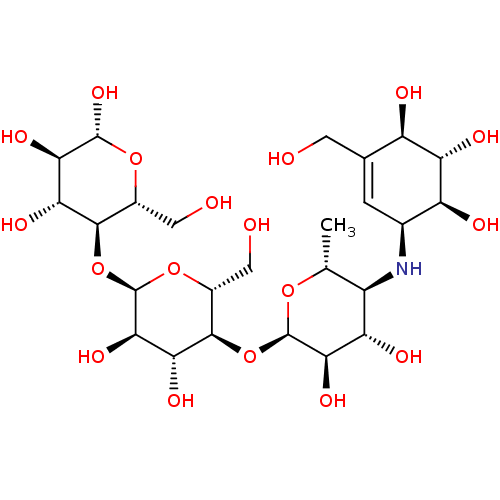
Report error Found 22 Enz. Inhib. hit(s) with all data for entry = 6086
Affinity DataIC50: 5.50E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 5.80E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 6.50E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 6.80E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 7.20E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 7.50E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 8.20E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 8.50E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 8.60E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 8.70E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair
Affinity DataIC50: 8.51E+4nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 9.08E+4nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 9.82E+4nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 1.13E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 1.98E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 2.05E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 2.21E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 2.30E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 2.46E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 2.51E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 2.60E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair