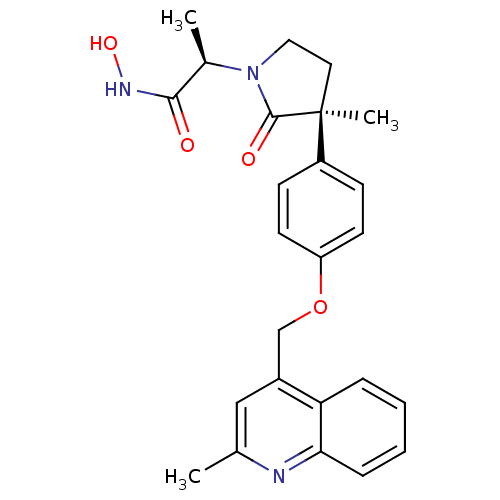
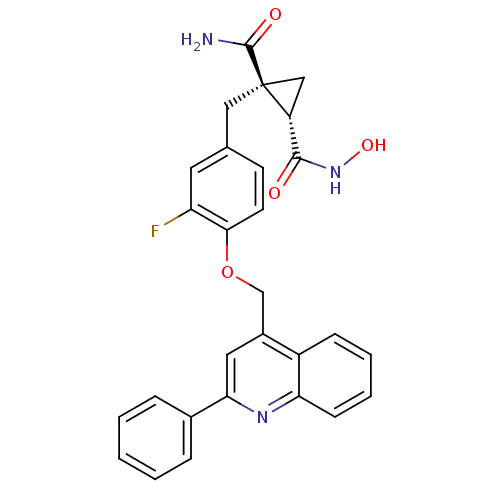
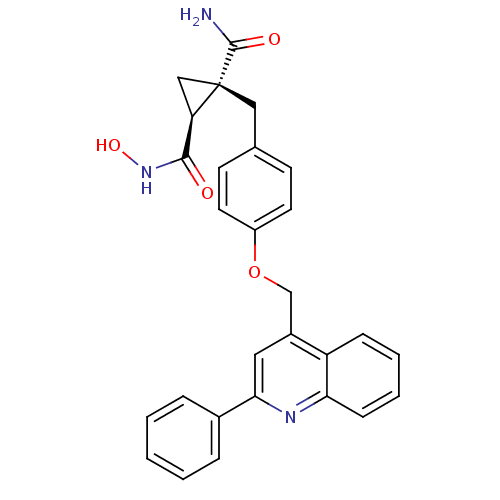
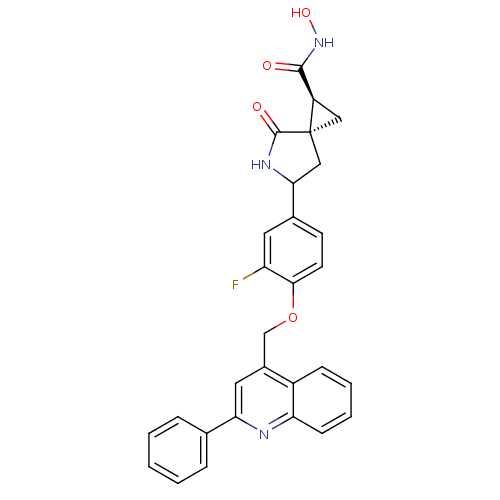
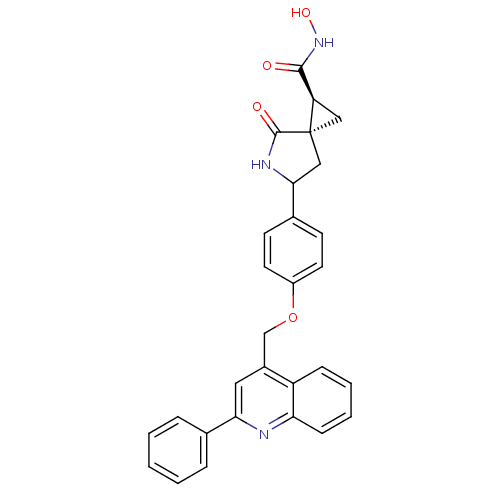
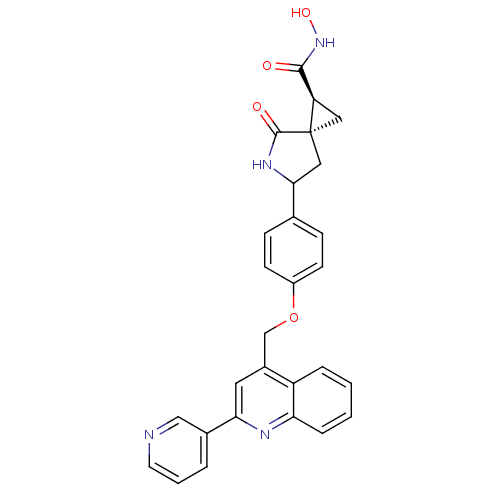
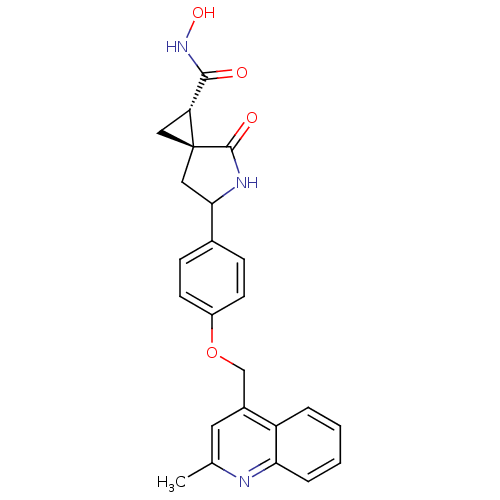
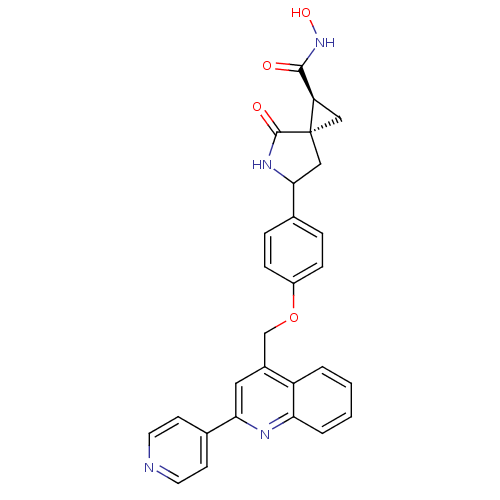
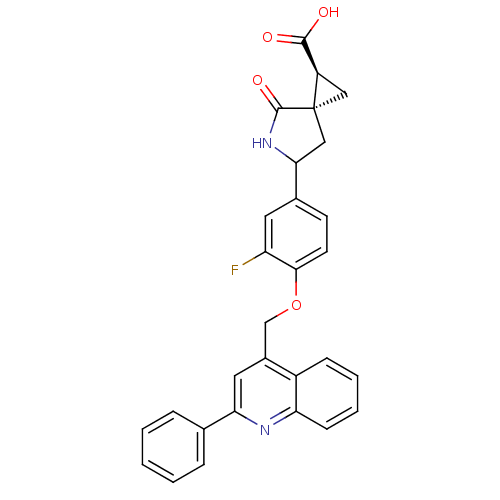
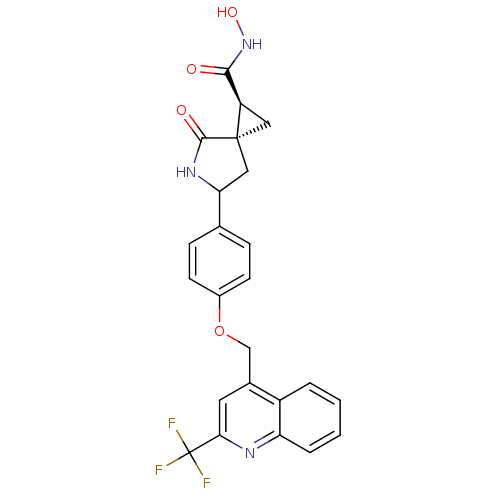
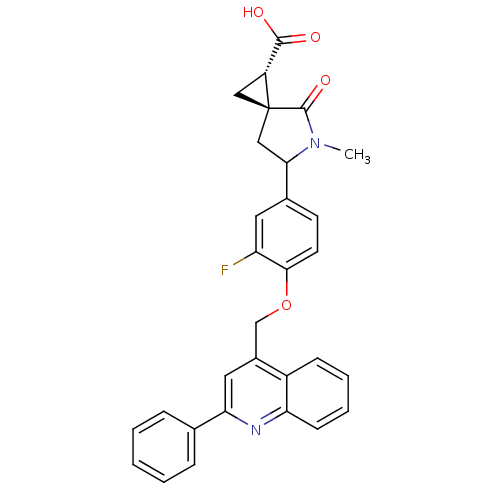
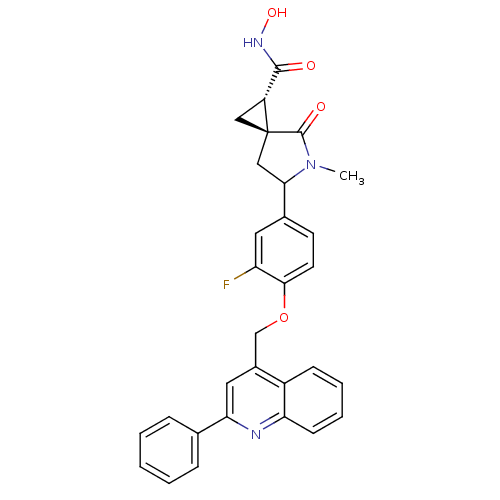
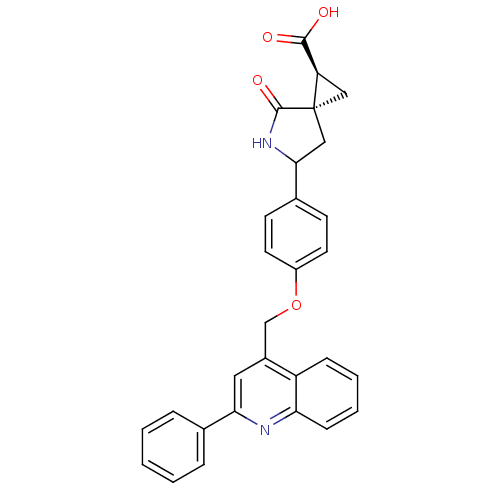
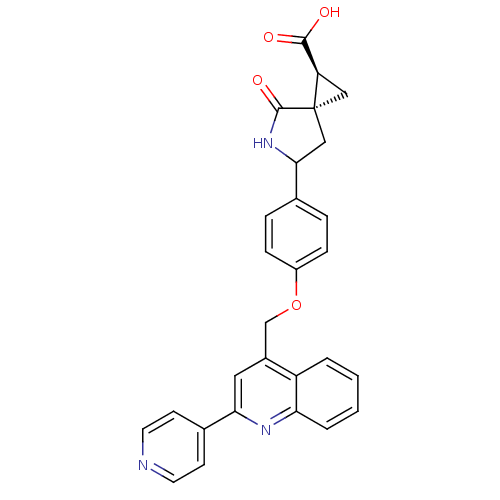
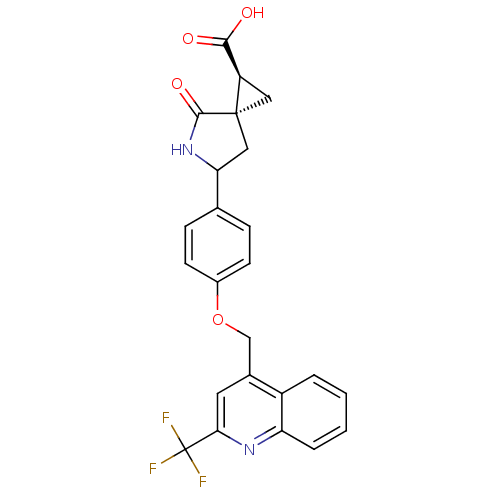
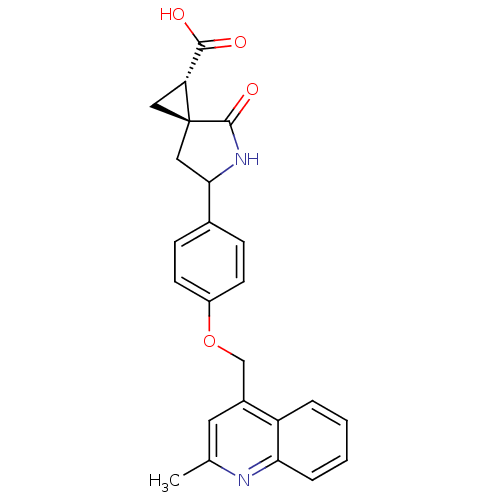
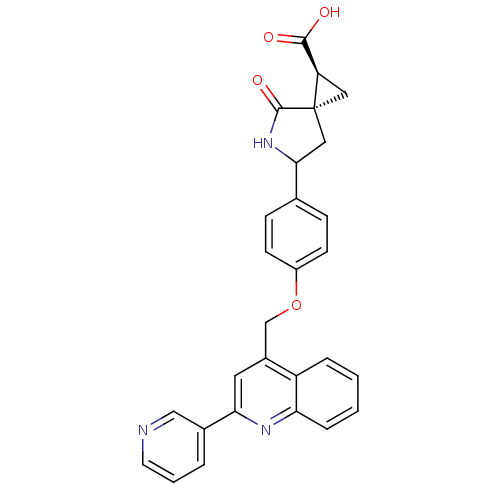
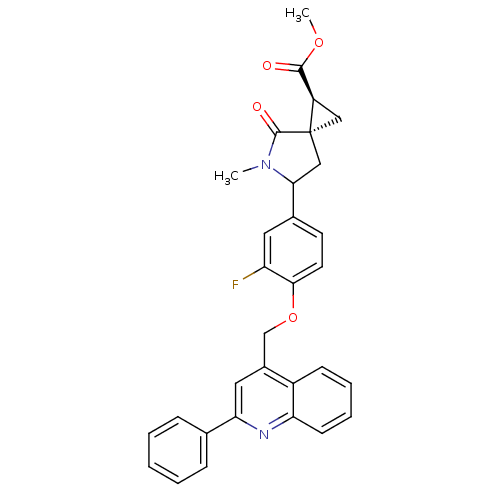
Report error Found 19 Enz. Inhib. hit(s) with all data for entry = 2977
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 0.0600nM ΔG°: -57.8kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 0.140nM ΔG°: -55.7kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 0.180nM ΔG°: -55.1kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 3nM ΔG°: -48.2kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 6.70nM ΔG°: -46.2kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 30nM ΔG°: -42.5kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 32nM ΔG°: -42.3kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 32nM ΔG°: -42.3kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 33nM ΔG°: -42.3kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 40nM ΔG°: -41.8kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 53nM ΔG°: -41.1kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 94nM ΔG°: -39.7kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 137nM ΔG°: -38.8kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 143nM ΔG°: -38.7kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 673nM ΔG°: -34.9kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 2.14E+3nM ΔG°: -32.0kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 3.07E+3nM ΔG°: -31.1kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 3.22E+3nM ΔG°: -31.0kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair
TargetDisintegrin and metalloproteinase domain-containing protein 17 [215-477,S266A,N452Q](Human)
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: >5.00E+4nM ΔG°: >-24.3kJ/molepH: 7.3 T: 2°CAssay Description:Enzyme activity was determined by a kinetic assay measuring the rate of increase in fluorescent intensity generated by the cleavage of an internally ...More data for this Ligand-Target Pair