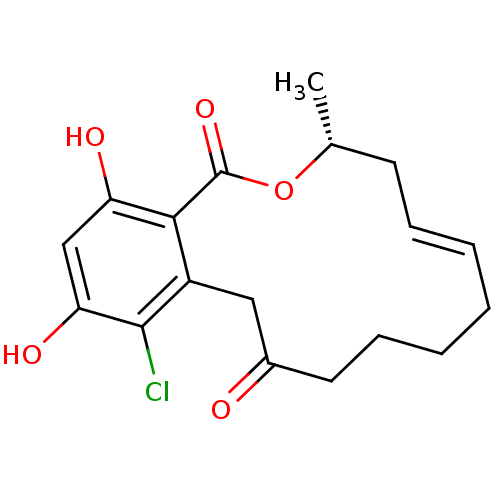
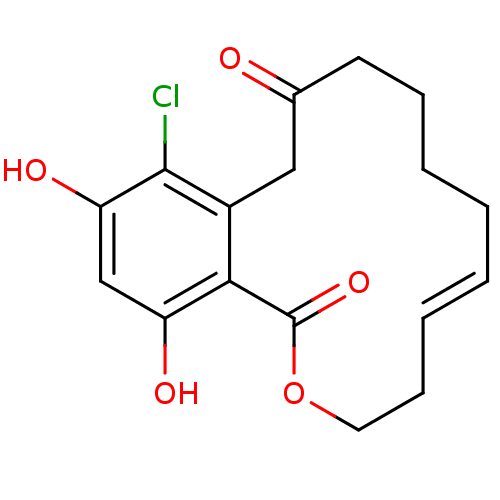
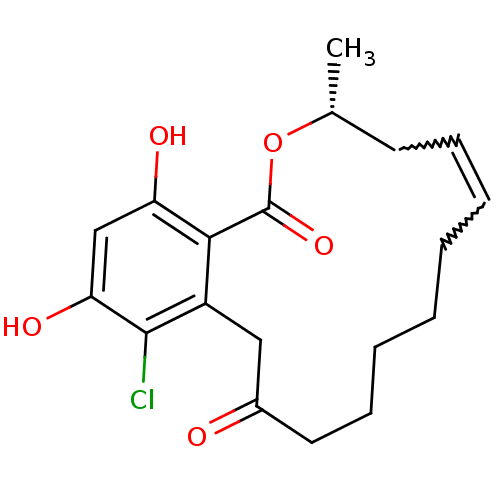
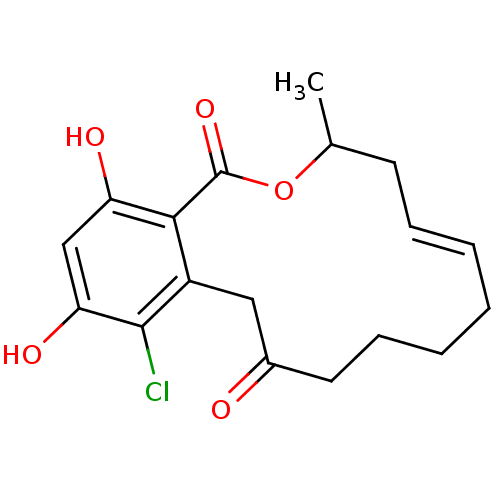
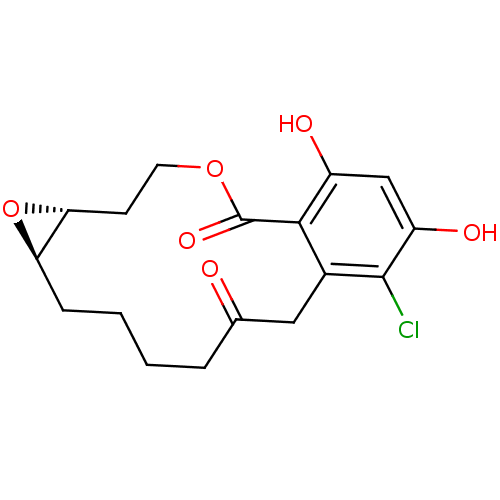
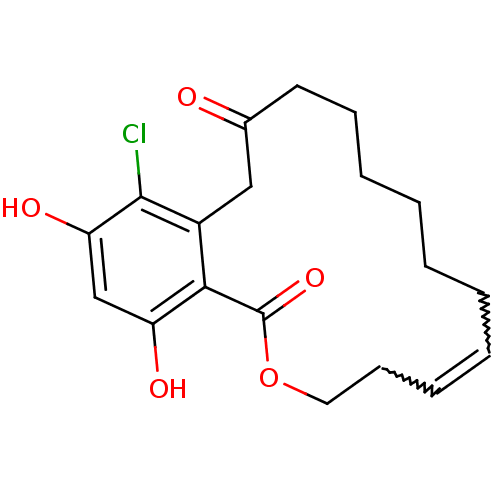
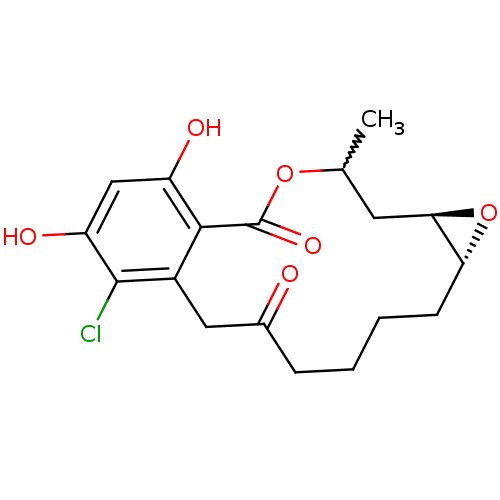
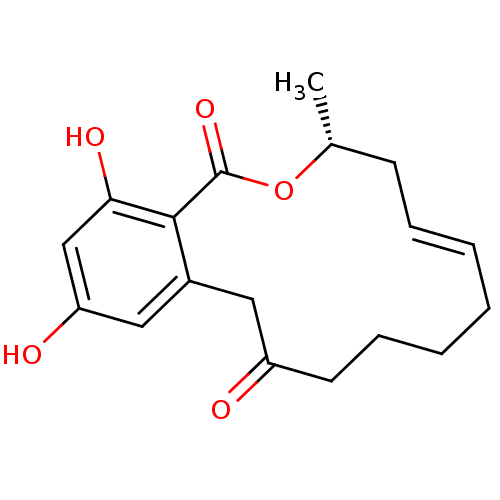
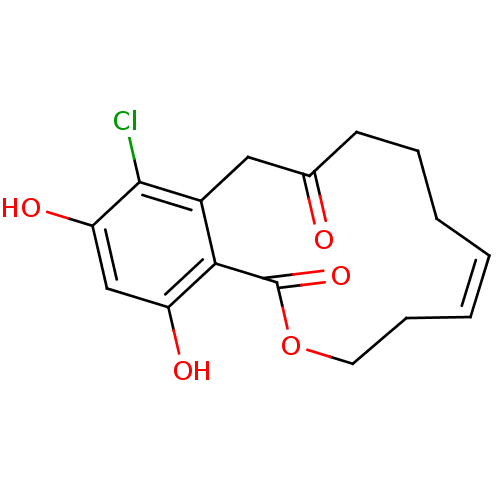
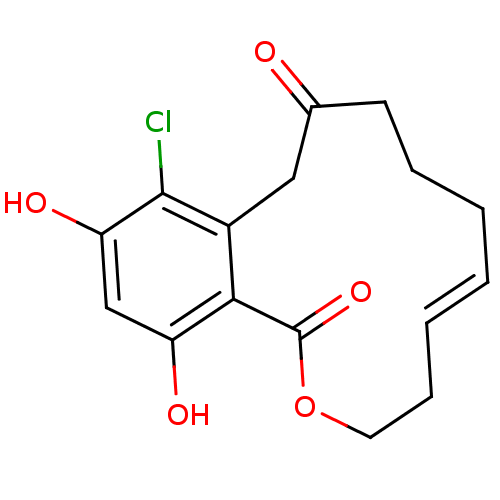
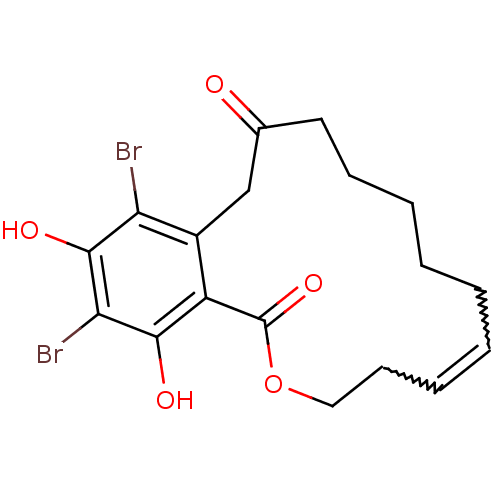
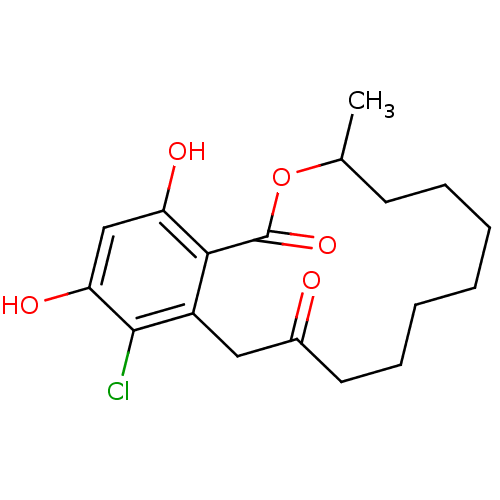
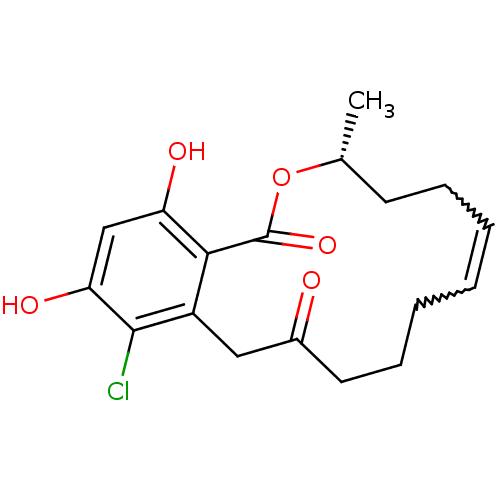
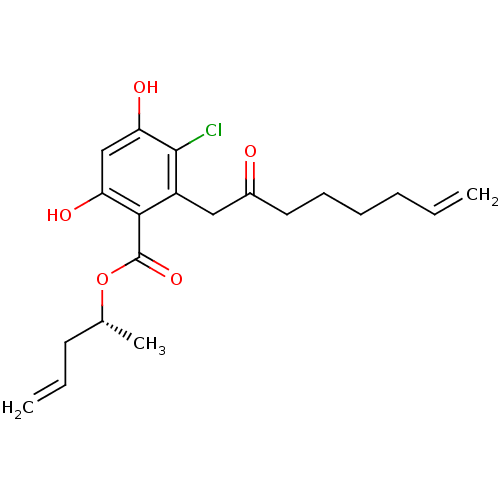
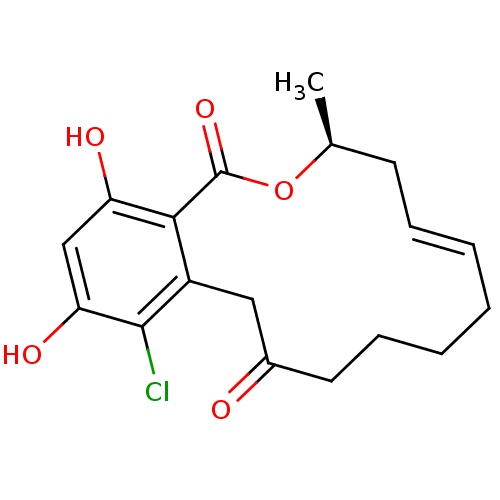
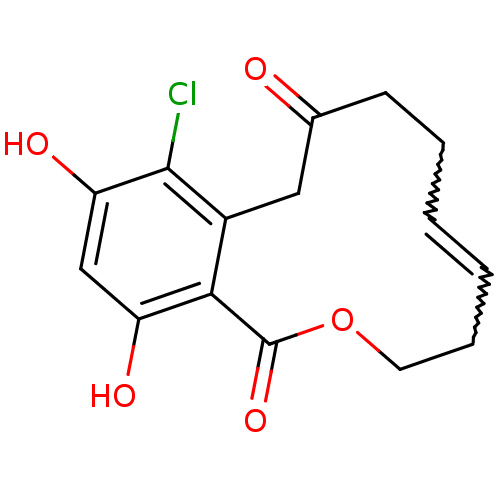
Report error Found 63 Enz. Inhib. hit(s) with all data for entry = 4411
Affinity DataIC50: 30nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 47nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 100nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 100nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 100nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 120nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 160nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 160nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 160nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 190nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 220nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 240nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 250nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 310nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 310nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 310nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 500nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 500nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 540nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 590nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 760nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 850nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 870nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 1.10E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 1.29E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 2.40E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 2.63E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 2.90E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 3.20E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 3.23E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 3.40E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 3.50E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 3.80E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 4.00E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 4.18E+3nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 4.30E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 4.90E+3nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
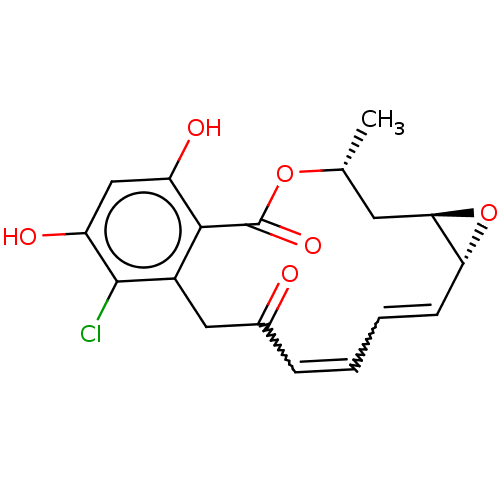
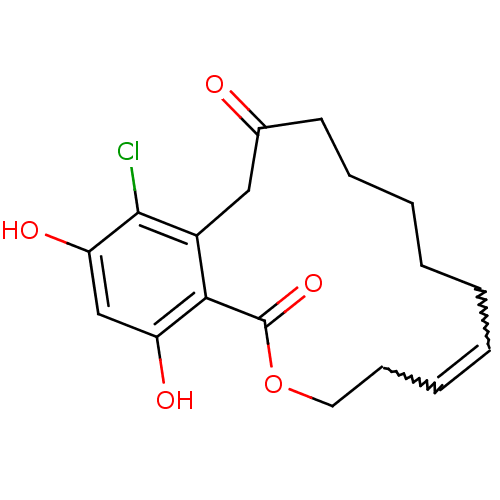
 3D Structure (crystal)
3D Structure (crystal)