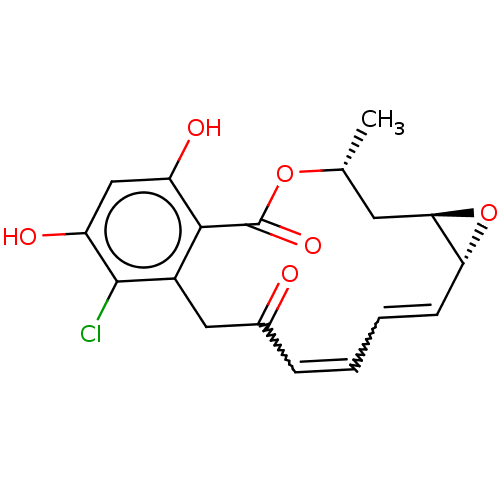
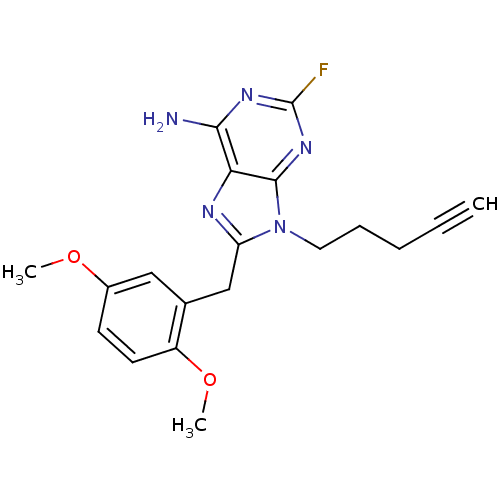
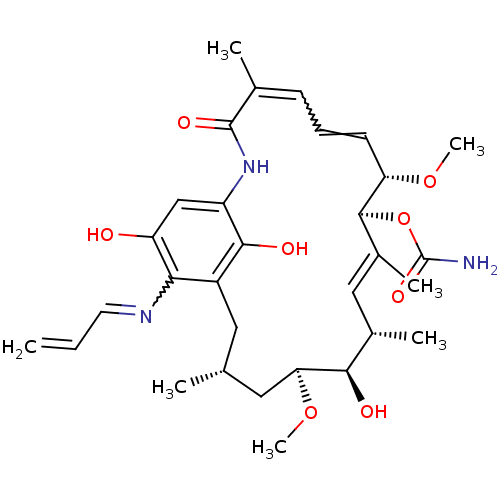
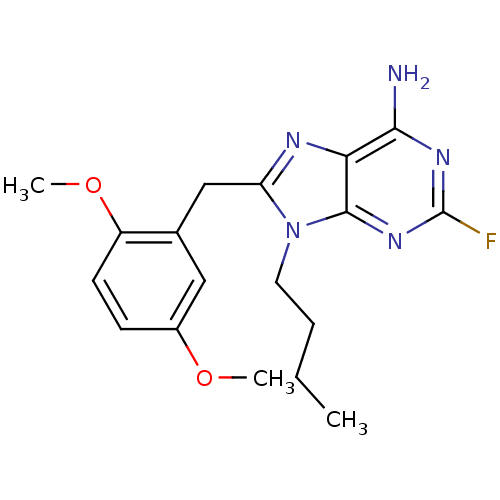
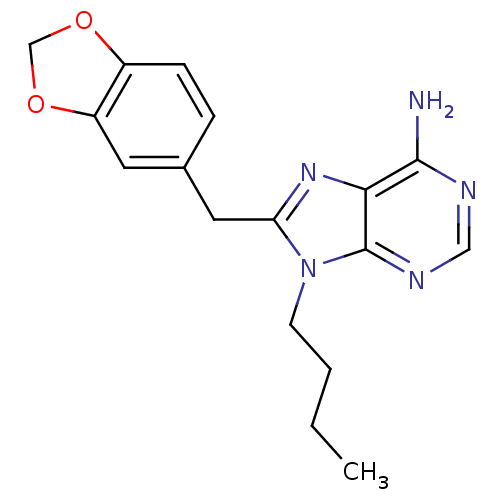
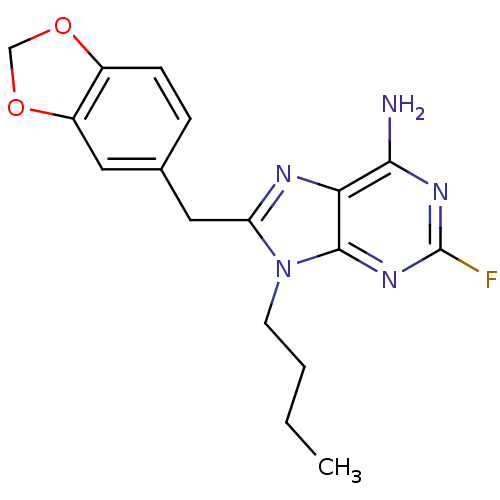
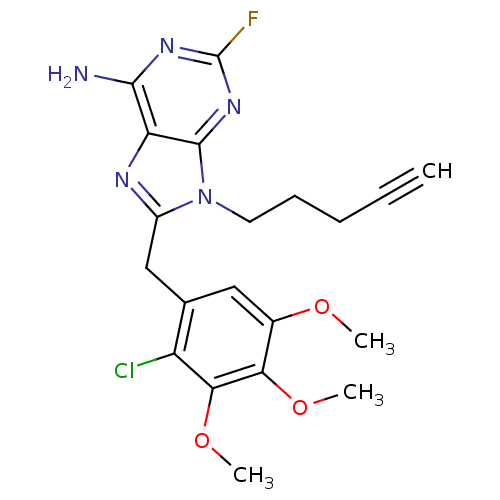
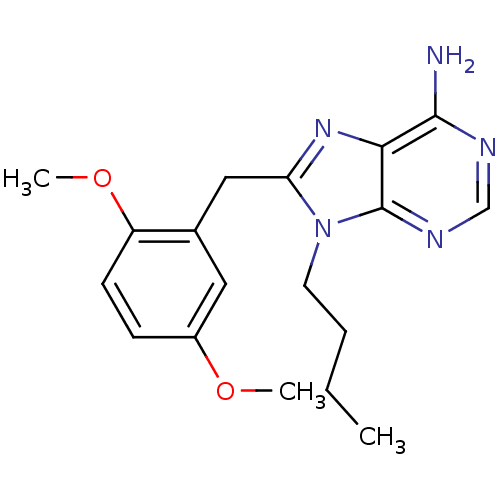
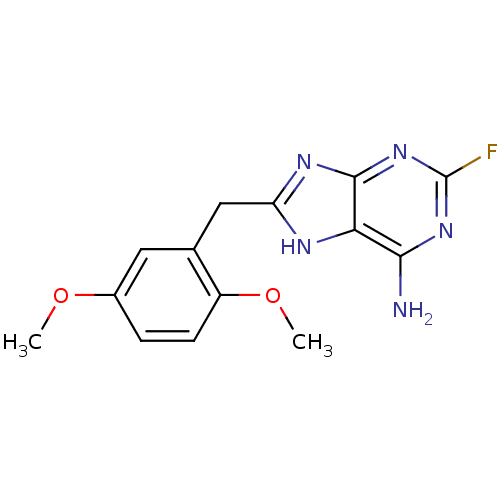
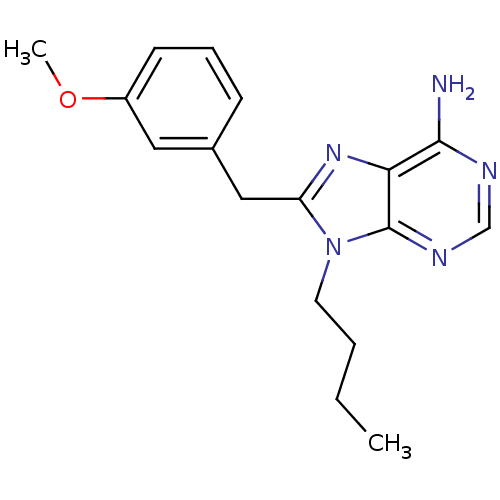
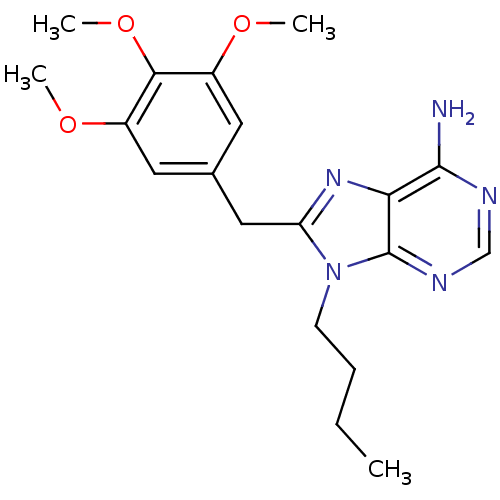
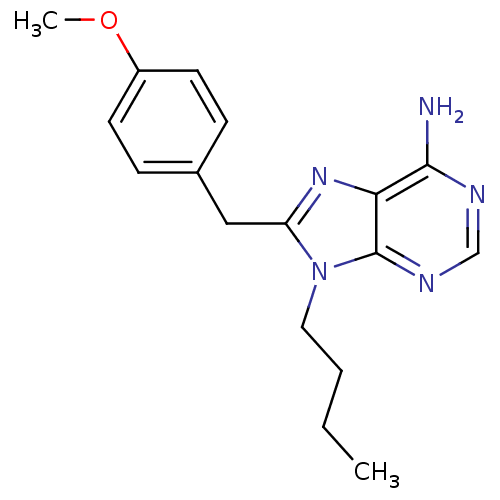
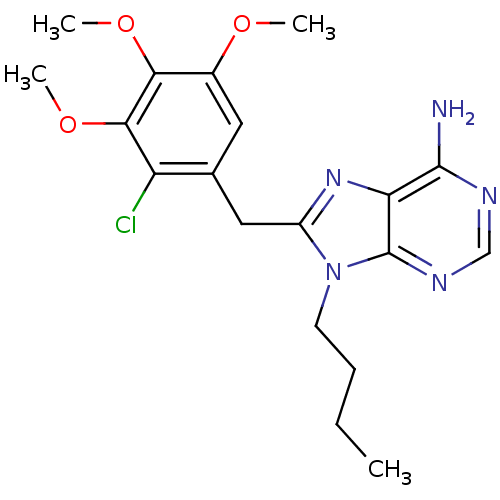
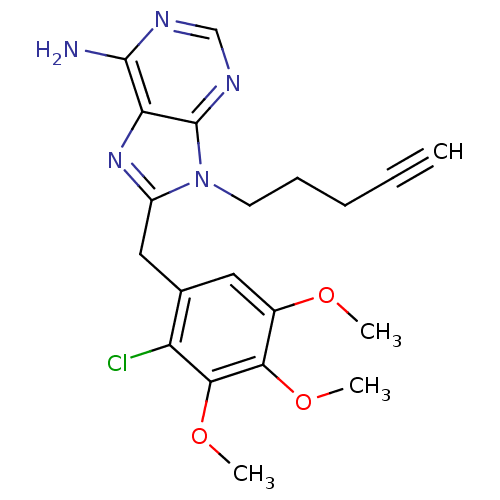
Report error Found 14 Enz. Inhib. hit(s) with all data for entry = 1948
Affinity DataIC50: 200nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 4.10E+3nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 1.34E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 1.43E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 1.53E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 1.71E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 4.10E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 5.35E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 7.50E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair