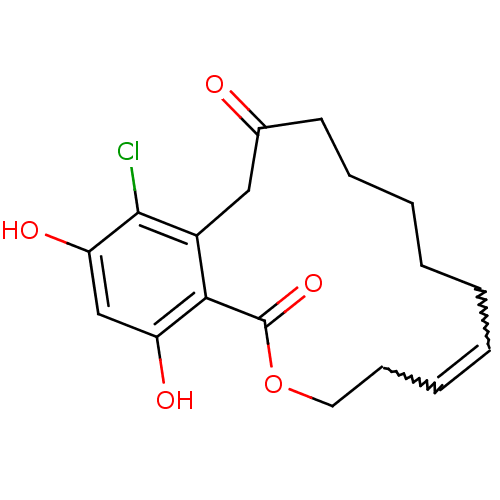
BDBM68263 Microlactone, 15h
SMILES c1c(c2c(c(c1O)Cl)CC(=O)CCCCC\C=C\CCOC2=O)O
InChI Key InChIKey=VZTAZMSAAIUZJV-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 68263
Found 3 hits for monomerid = 68263
Affinity DataIC50: 100nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 160nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 760nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)