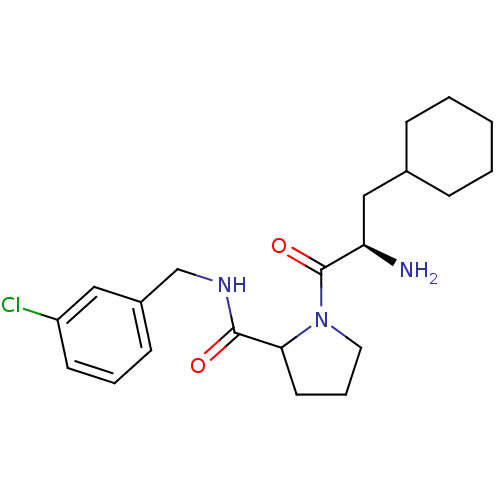
BDBM32658 m-chlorobenzylamide, 7
SMILES N[C@H](CC1CCCCC1)C(=O)N1CCCC1C(=O)NCc1cccc(Cl)c1
InChI Key InChIKey=JGFCNVHEEMBVJG-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 32658
Found 2 hits for monomerid = 32658
Affinity DataKi: 100nM ΔG°: -9.54kcal/molepH: 7.4 T: 2°CAssay Description:Kinetic inhibition of human thrombin was determined photometrically at 405 nm using the chromogenic substrate Pefachrom tPa. Reactions were performed...More data for this Ligand-Target Pair
Affinity DataKi: 100nMAssay Description:Inhibition of human thrombin using pefachrom tPa as substrate after 3 mins by photometric methodMore data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 4 hits for monomerid = 32658
Found 4 hits for monomerid = 32658
ITC DataΔG°: -9.29kcal/mole −TΔS°: -0.812kcal/mole ΔH°: -8.50kcal/mole
pH: 7.8 T: 25.00°C
pH: 7.8 T: 25.00°C
ITC DataΔG°: -8.69kcal/mole −TΔS°: -1.00kcal/mole ΔH°: -7.69kcal/mole
pH: 7.8 T: 25.00°C
pH: 7.8 T: 25.00°C
ITC DataΔG°: -9.48kcal/mole −TΔS°: -1.79kcal/mole ΔH°: -7.67kcal/mole
pH: 7.8 T: 25.00°C
pH: 7.8 T: 25.00°C
ITC DataΔG°: -9.34kcal/mole −TΔS°: -1.79kcal/mole ΔH°: -7.55kcal/mole
pH: 7.8 T: 25.00°C
pH: 7.8 T: 25.00°C