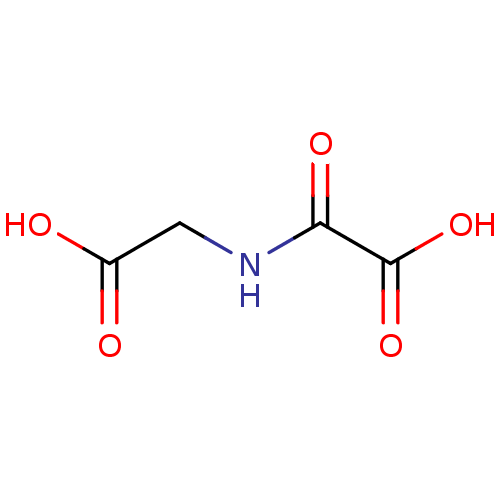
BDBM26106 CHEMBL90852::N-oxalyl glycine, 1a::NOG::Oxalylglycine::[(carboxymethyl)carbamoyl]formic acid
SMILES C(C(=O)O)NC(=O)C(=O)O
InChI Key InChIKey=BIMZLRFONYSTPT-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 63 hits for monomerid = 26106
Found 63 hits for monomerid = 26106
Affinity DataIC50: 8.00E+3nMAssay Description:Inhibition of recombinant human PHD2 using 2OG as substrate and Fe2 as co-factor assessed as hydroxylation incubated for 15 mins in presence of L-asc...More data for this Ligand-Target Pair
Affinity DataIC50: 7.80E+4nMpH: 7.5 T: 2°CAssay Description:A coupled-assay for JMJD2E activity employing formaldehyde dehydrogenase (FDH) from Pseudomonas putida was developed. Formaldehyde release by demethy...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 6.40E+5nMAssay Description:Inhibition of KDM5A (unknown origin) using H3K4me3 peptide and 2-oxoglutarate as substrate after 1 hr by FDH-coupled assayMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 4C(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 2.50E+5nMAssay Description:Inhibition of KDM4C (unknown origin) using H3K9me3 peptide and 2-oxoglutarate as substrate after 1 hr by FDH-coupled assayMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 4A(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 2.50E+5nMAssay Description:Inhibition of KDM4A (unknown origin) using H3K9me3 peptide and 2-oxoglutarate as substrate after 1 hr by FDH-coupled assayMore data for this Ligand-Target Pair
Affinity DataIC50: 4.40E+4nMAssay Description:Inhibition of human hexahistidine-tagged full-length FTO expressed in Escherichia coli BL21 (DE3) using 3-methylthymidine as substrate assessed as in...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+4nMAssay Description:Inhibition of human PHD2 catalytic domain (181 to 426) Mn2 expressed in Escherichia coli by NMR spectroscopic analysisMore data for this Ligand-Target Pair
Affinity DataKd: 3.00E+3nMAssay Description:Binding affinity to human PHD2 catalytic domain (181 to 426) Mn2 expressed in Escherichia coli by NMR spectroscopic analysisMore data for this Ligand-Target Pair
Affinity DataKd: 2.00E+3nMAssay Description:Displacement of [13C]-2OG from catalytic domain of PHD2 (181 to 426) (unknown origin) expressed in Escherichia coliMore data for this Ligand-Target Pair
Affinity DataIC50: 5.01E+4nMAssay Description:Inhibition of KDM2A (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 316nMAssay Description:Inhibition of KDM6B (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:Inhibition of KDM3A (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 7.94E+3nMAssay Description:Inhibition of KDM5C (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of KDM4E (unknown origin)More data for this Ligand-Target Pair
TargetLysine-specific demethylase 4A(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 8.50E+4nMAssay Description:Inhibition of C-terminal FLAG-tagged human KDM4A expressed in Escherichia coli pre-incubated for 4 hrs before substrate addition and measured after 1...More data for this Ligand-Target Pair
Affinity DataIC50: 2.12E+4nMAssay Description:Inhibition of N-terminal His6-tagged recombinant Paramecium bursaria chlorella virus 1 CPH expressed in Escherichia coli Rosetta 2 (DE3) cells pre-in...More data for this Ligand-Target Pair
Affinity DataIC50: 4.50E+4nMAssay Description:Inhibition of KDM2A (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 1.97E+4nMAssay Description:Inhibition of recombinant human GST-tagged TET2 (1099 to 1936 residues) expressed in Escherichia coli BL21 (DE3)pLysS assessed as reduction in 5hmc l...More data for this Ligand-Target Pair
Affinity DataIC50: 2.45E+4nMAssay Description:Inhibition of KDM5B (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 1.13E+4nMAssay Description:Inhibition of N-terminal His6-tagged TET2 (unknown origin) expressed in Escherichia coli assessed as reduction in 5-methylcytosine oxidation by measu...More data for this Ligand-Target Pair
TargetHypoxia-inducible factor 1-alpha inhibitor(Human)
China Pharmaceutical University
Curated by ChEMBL
China Pharmaceutical University
Curated by ChEMBL
Affinity DataIC50: 4.60E+4nMAssay Description:Inhibition of FIH (unknown origin) assessed as hydroxylation of HIF1alphaCAD by MALDI-TOF-MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 9.20E+5nMAssay Description:Inhibition of KDM5B (unknown origin)More data for this Ligand-Target Pair
TargetAlpha-ketoglutarate-dependent dioxygenase AlkB(Escherichia coli (strain K12))
University College London
Curated by ChEMBL
University College London
Curated by ChEMBL
Affinity DataIC50: 1.08E+5nMAssay Description:Inhibition of Escherichia coli AlkB using 15-mer m1A-ssDNA as substrate incubated for 10 mins by HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+4nMAssay Description:Inhibition of N-terminal 3xFlag-tagged human TET1 catalytic domain (E1418 to V2136 residues) expressed in Sf9 cells using 5-methylcytosine as substra...More data for this Ligand-Target Pair
Affinity DataIC50: 9.00E+3nMAssay Description:Inhibition of His10-FLAG tagged human TET2 catalytic domain (Q969 to I2002 residues) expressed in Sf9 cells using 5-methylcytosine as substrate prein...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+3nMAssay Description:Inhibition of human TET3 catalytic domain (E824 to I1795 residues) expressed in mammalian cells using 5-methylcytosine as substrate preincubated for ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.65E+4nMAssay Description:Inhibition of human full length FTO expressed in Escherichia coli BL21 (DE3) Rosetta T1R cells using 3-methylthymidine as substrate incubated for 1 h...More data for this Ligand-Target Pair
Affinity DataIC50: 1.25E+5nMAssay Description:Inhibition of human ALKBH2 (56 to 258 residues) expressed in Escherichia coli BL21 (DE3) cells using 15-mer m1A-ssDNA as substrate incubated for 10 m...More data for this Ligand-Target Pair
TargetAlpha-ketoglutarate-dependent dioxygenase alkB homolog 3(Human)
University College London
Curated by ChEMBL
University College London
Curated by ChEMBL
Affinity DataIC50: 9.68E+4nMAssay Description:Inhibition of N-terminal His-tagged human ALKBH3 (1 to 286 residues) using m6A-ssDNA as substrate incubated for 30 mins by MALDI-TOF-MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.59E+4nMAssay Description:Inhibition of ALKBH5 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 2.45E+4nMAssay Description:Inhibition of KDM5B (unknown origin)More data for this Ligand-Target Pair
Affinity DataKd: 3.10E+3nMAssay Description:Binding affinity to human PHD2 by nondenaturing ESI-MSMore data for this Ligand-Target Pair
Affinity DataIC50: 6.90E+3nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 2.45E+4nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 3.50E+3nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 5.80E+3nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 4C(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 4.15E+4nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 680nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 290nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 4.59E+4nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 2.89E+3nMAssay Description:Compound was evaluated for the inhibition of prolyl 4-hydroxylaseMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 4A(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 2.50E+5nMAssay Description:Inhibition of recombinant His-tagged JMJD2A expressed in Escherichia coli Rosetta 2 (DE3) pLysS cells by formaldehyde dehydrogenase-coupled assayMore data for this Ligand-Target Pair
TargetHistone lysine demethylase PHF8(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 6.40E+5nMAssay Description:Inhibition of JHDM1F (unknown origin) using H3K9mel peptideMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 2.50E+5nMAssay Description:Inhibition of JARID1A (unknown origin) using biotinylated H3K9mel peptide after 60 mins by Alpha screen assayMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 4C(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 4.30E+5nMAssay Description:Inhibition of JMJD2C (unknown origin) using H3K9mel peptideMore data for this Ligand-Target Pair
Affinity DataIC50: 5.69E+5nMAssay Description:Inhibition of JMJD1A (unknown origin) using H3K9mel peptideMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 4A(Human)
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Kyoto Prefectural University of Medicine
Curated by ChEMBL
Affinity DataIC50: 2.50E+5nMAssay Description:Inhibition of KDM4AMore data for this Ligand-Target Pair
Affinity DataIC50: 1.85E+4nMAssay Description:Inhibition of human PHD2 at 293K temperature by solvent relaxation techniqueMore data for this Ligand-Target Pair
Affinity DataKd: 2.80E+3nMAssay Description:Binding affinity to human PHD2-Mn(II) using 100% H2O MQC spectrometer operated at 23 MHz at 313K temperatureMore data for this Ligand-Target Pair
Affinity DataKd: 3.10E+3nMAssay Description:Binding affinity to human PHD2-Mn(II) using 12.5% H2O/87.5% D2O MQC spectrometer operated at 500 MHz at 298K temperatureMore data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)