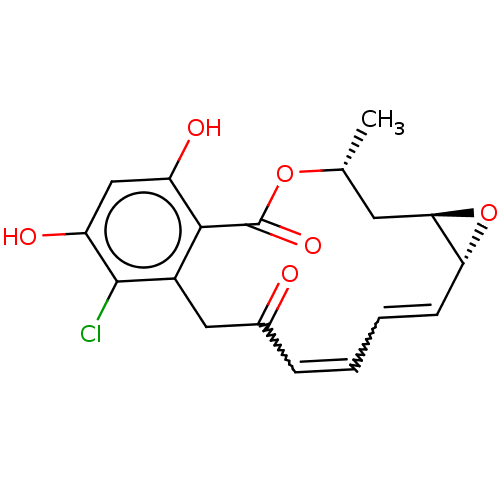
BDBM15361 (1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-methyl-1a,14,15,15a-tetrahydro-6H-oxireno[e][2]benzoxacyclotetradecine-6,12(7H)-dione::(4R,6R,8R,9Z,11E)-16-chloro-17,19-dihydroxy-4-methyl-3,7-dioxatricyclo[13.4.0.0^{6,8}]nonadeca-1(19),9,11,15,17-pentaene-2,13-dione::CHEMBL414883::Microlactone, 1::Monorden::Radicicol
SMILES C[C@@H]1C[C@H]2O[C@@H]2C=CC=CC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1
InChI Key InChIKey=WYZWZEOGROVVHK-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 8 hits for monomerid = 15361
Found 8 hits for monomerid = 15361
Affinity DataIC50: 150nMpH: 7.4 T: 2°CAssay Description:The assay is based upon displacement of a fluorescently labeled molecule, which binds specifically to the ATP-binding site of full-length human Hsp90...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
Affinity DataIC50: 150nMpH: 7.4 T: 2°CAssay Description:The assay is based upon displacement of a fluorescently labeled molecule, which binds specifically to the ATP-binding site of full-length human Hsp90...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 47nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataIC50: 30nMAssay Description:Binding competition assay with a fluorescent probe.More data for this Ligand-Target Pair
Affinity DataKi: 7.00E+3nMAssay Description:Competitive inhibition of rat liver ACLY using ATP as substrate by spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.30E+4nMAssay Description:Competitive inhibition of rat liver ACLY using citrate as substrate by spectrophotometric analysisMore data for this Ligand-Target Pair