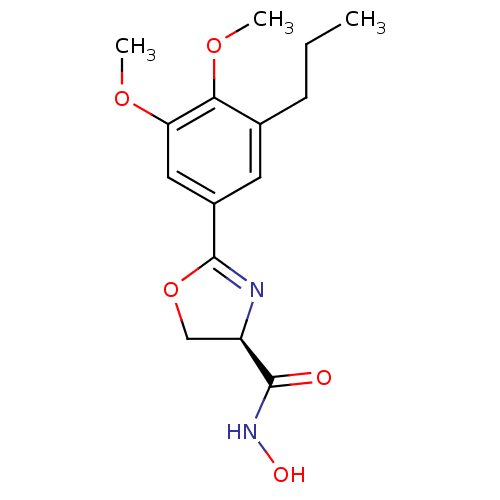
BDBM50074960 (R)-2-(3,4-Dimethoxy-5-propyl-phenyl)-4,5-dihydro-oxazole-4-carboxylic acid hydroxyamide::(R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydrooxazole-4-carboxamide::CHEMBL107313::L-161240
SMILES CCCc1cc(cc(c1OC)OC)C2=N[C@H](CO2)C(=O)NO
InChI Key InChIKey=DGGKRIGJIQRXQD-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 7 hits for monomerid = 50074960
Found 7 hits for monomerid = 50074960
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Escherichia coli (strain K12))
Merck Research Laboratories
Curated by ChEMBL
Merck Research Laboratories
Curated by ChEMBL
Affinity DataIC50: 30nMAssay Description:Inhibition of UDP-3-O-[R-3-hydroxymyristoyl]-GlcNAc deacetylase using direct deacetylase assay (DEACET) in Escherichia coli strain JB 1104More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Escherichia coli (strain K12))
Merck Research Laboratories
Curated by ChEMBL
Merck Research Laboratories
Curated by ChEMBL
Affinity DataKi: 0.0530nMAssay Description:Inhibition of Escherichia coli LpxCMore data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Escherichia coli (strain K12))
Merck Research Laboratories
Curated by ChEMBL
Merck Research Laboratories
Curated by ChEMBL
Affinity DataKi: 0.340nMAssay Description:Inhibition of Escherichia coli LpxC Q202W/G210S mutantMore data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Escherichia coli (strain K12))
Merck Research Laboratories
Curated by ChEMBL
Merck Research Laboratories
Curated by ChEMBL
Affinity DataKi: 50nMAssay Description:Inhibition of Escherichia coli LpxCMore data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Escherichia coli (strain K12))
Merck Research Laboratories
Curated by ChEMBL
Merck Research Laboratories
Curated by ChEMBL
Affinity DataKi: 50nMAssay Description:Inhibition of Escherichia coli LpxCMore data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Escherichia coli (strain K12))
Merck Research Laboratories
Curated by ChEMBL
Merck Research Laboratories
Curated by ChEMBL
Affinity DataKi: 50nMAssay Description:Inhibition of Escherichia coli LpxC enzymeMore data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Rhizobium leguminosarum bv. trifolii (strain WSM13...)
Duke University Medical Center
Curated by ChEMBL
Duke University Medical Center
Curated by ChEMBL
Affinity DataKi: 110nMAssay Description:Inhibition of Rhizobium leguminosarum LpxC W206Q/S214G mutantMore data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)