BDBM50646965 CHEMBL5595943
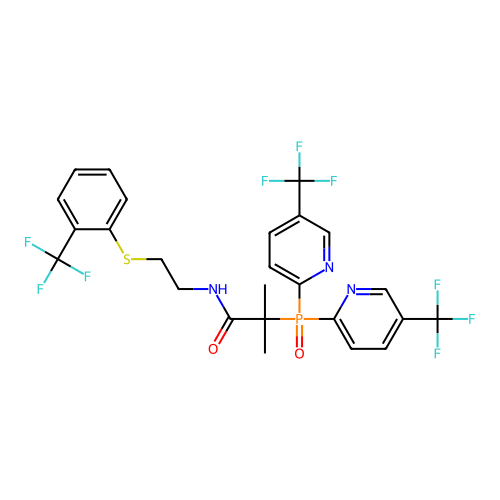
SMILES CC(C)(C(=O)NCCSc1ccccc1C(F)(F)F)P(=O)(c1ccc(C(F)(F)F)cn1)c1ccc(C(F)(F)F)cn1
InChI Key
PDB links: 2 PDB IDs match this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50646965
Found 4 hits for monomerid = 50646965
TargetATP-dependent Clp protease proteolytic subunit(Neisseria gonorrhoeae (strain ATCC 700825 / FA 109...)
University of Toronto
Curated by ChEMBL
University of Toronto
Curated by ChEMBL
Affinity DataEC50: 160nMAssay Description:Activation of N-terminal His-tagged recombinant Neisseria gonorrhoeae ClpP expressed in Escherichia coli SG1146(DE3) assessed as degradation of casei...More data for this Ligand-Target Pair
TargetATP-dependent Clp protease proteolytic subunit(Escherichia coli (strain K12))
University of Toronto
Curated by ChEMBL
University of Toronto
Curated by ChEMBL
Affinity DataEC50: 200nMAssay Description:Activation of N-terminal His-tagged recombinant Escherichia coli ClpP expressed in Escherichia coli SG1146(DE3) assessed as degradation of casein-FIT...More data for this Ligand-Target Pair
TargetATP-dependent Clp protease proteolytic subunit(Escherichia coli (strain K12))
University of Toronto
Curated by ChEMBL
University of Toronto
Curated by ChEMBL
Affinity DataKd: 270nMAssay Description:Binding affinity to His-tagged recombinant Escherichia coli ClpP extracted from Escherichia coli SG1146(DE3) using casein as substrate assessed as ap...More data for this Ligand-Target Pair
TargetATP-dependent Clp protease proteolytic subunit(Neisseria gonorrhoeae (strain ATCC 700825 / FA 109...)
University of Toronto
Curated by ChEMBL
University of Toronto
Curated by ChEMBL
Affinity DataKd: 690nMAssay Description:Binding affinity to His-tagged recombinant Neisseria gonorrhoeae ClpP extracted from Escherichia coli SG1146(DE3) using casein as substrate assessed ...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)