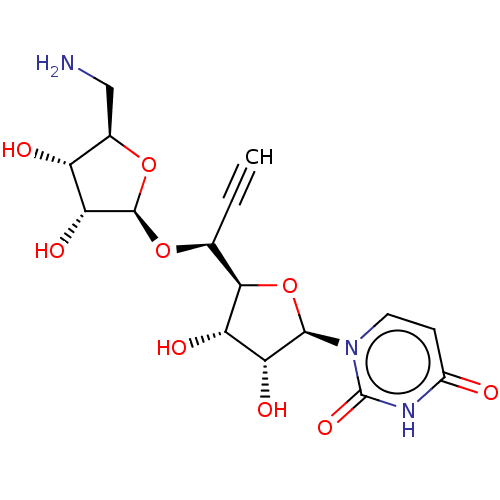
BDBM50526649 CHEMBL4556639
SMILES [H][C@@]1(O[C@@H](C#C)[C@@]2([H])O[C@H]([C@H](O)[C@@H]2O)n2ccc(=O)[nH]c2=O)O[C@H](CN)[C@@H](O)[C@H]1O
InChI Key InChIKey=ZMPJDHGJNDQVLQ-UHFFFAOYSA-N
Data 5 IC50
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 5 hits for monomerid = 50526649
Found 5 hits for monomerid = 50526649
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis (strain 168))
Griffith University
Curated by ChEMBL
Griffith University
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of MraY in Bacillus subtilisMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Staphylococcus aureus)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataIC50: 2.00E+4nMAssay Description:Inhibition of Staphylococcus aureus MraY assessed as reduction of dansylated lipid I formation using UDP-MurNAcdansylpentapeptide as substrate incuba...More data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis (strain 168))
Griffith University
Curated by ChEMBL
Griffith University
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of Bacillus subtilis MraY using UDP-MurNAc-pentapeptide as substrate incubated for 30 mins by radioactivity based analysisMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Staphylococcus aureus)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataIC50: 9.00E+4nMAssay Description:Inhibition of Staphylococcus aureus MraY using UDP-MurNAc-dansylpentapeptide as substrate incubated for 3 hrs by fluorescence based microplate reader...More data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis (strain 168))
Griffith University
Curated by ChEMBL
Griffith University
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of Bacillus subtilis MraYMore data for this Ligand-Target Pair