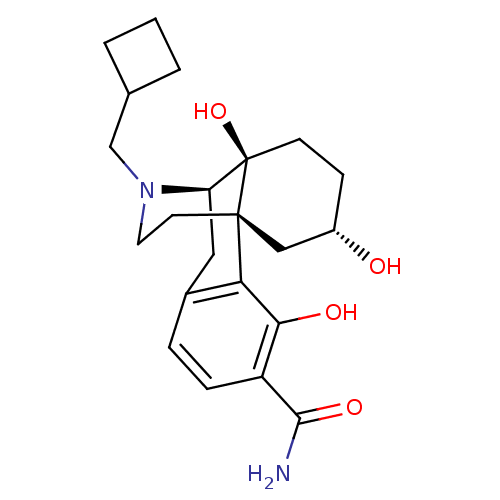
BDBM50278335 (1R,9R,10S,13S)-17-(cyclobutylmethyl)-3,10,13-trihydroxy-17-azatetracyclo[7.5.3.0^{1,10}.0^{2,7}]heptadeca-2(7),3,5-triene-4-carboxamide::CHEMBL512150::US10231963, Table B.19::US10287250, Compound B.13::US10736890, Compound TABLE B.19::US10752592, Compound TABLE B.13::US11534436, Compound Table B.19::US9133125, Table B, Compound 13::US9656961, Example 00136
SMILES NC(=O)c1ccc2C[C@H]3N(CC4CCC4)CC[C@@]4(C[C@@H](O)CC[C@@]34O)c2c1O
InChI Key InChIKey=NWIALXFYKPFWTR-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 24 hits for monomerid = 50278335
Found 24 hits for monomerid = 50278335
Affinity DataEC50: 2.80nMAssay Description:The EC50 and Imax for μ opioid receptors was determined using a [35S]GTPγS binding assay. This assay measures the functional properties of ...More data for this Ligand-Target Pair
Affinity DataIC50: 47nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataIC50: 47nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataEC50: 2.80nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataIC50: 7nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataIC50: 47nMAssay Description:The Ki (binding affinity) for opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Journal of ...More data for this Ligand-Target Pair
Affinity DataEC50: 2.80nMAssay Description:The Ki (binding affinity) for opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Journal of ...More data for this Ligand-Target Pair
Affinity DataIC50: 47nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataIC50: 47nMAssay Description:The Ki (binding affinity) for opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Journal of ...More data for this Ligand-Target Pair
Affinity DataEC50: 2.80nMAssay Description:The Ki (binding affinity) for opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Journal of ...More data for this Ligand-Target Pair
Affinity DataIC50: 26nMAssay Description:Antagonist activity against human mu opioid receptor expressed in CHO cells assessed as inhibition of DAMGO-stimulated [35S]GTPgammaS bindingMore data for this Ligand-Target Pair
Affinity DataEC50: 3.90nMAssay Description:Agonist activity at human kappa opioid receptor expressed in CHO cells assessed as stimulation of [35S]GTPgammaS bindingMore data for this Ligand-Target Pair
Affinity DataEC50: 3.10nMAssay Description:Agonist activity at human mu opioid receptor expressed in CHO cells assessed as stimulation of [35S]GTPgammaS bindingMore data for this Ligand-Target Pair
Affinity DataEC50: 2.80nMpH: 7.4Assay Description:The EC50 and Imax for mu opioid receptors was determined using a [I35S]GTPgammaS binding assay. This assay measures the functional properties of a co...More data for this Ligand-Target Pair
Affinity DataKi: 0.520nMAssay Description:Displacement of [3H]DAMGO from human mu opioid receptor expressed in CHO cells after 60 mins by scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 0.590nMAssay Description:The Ki (binding affinity) for opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Journal of ...More data for this Ligand-Target Pair
Affinity DataKi: 0.590nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataKi: 0.590nM ΔG°: -12.6kcal/mole IC50: 47nMpH: 7.5 T: 2°CAssay Description:The Ki (binding affinity) for u opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Journal o...More data for this Ligand-Target Pair
Affinity DataKi: 0.590nMAssay Description:The Ki (binding affinity) for opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Journal of ...More data for this Ligand-Target Pair
Affinity DataKi: 0.590nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataKi: 0.590nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataKi: 0.590nMAssay Description:The Ki (binding affinity) for μ opioid receptors was determined using a competitive displacement assay as previously described in Neumeyer (Jour...More data for this Ligand-Target Pair
Affinity DataKi: 9nMAssay Description:Displacement of [3H]U69,593 from human kappa opioid receptor expressed in CHO cells after 60 mins by scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 78nMAssay Description:Displacement of [3H]naltrindole from human delta opioid receptor expressed in CHO cells after 3 hrs by scintillation countingMore data for this Ligand-Target Pair