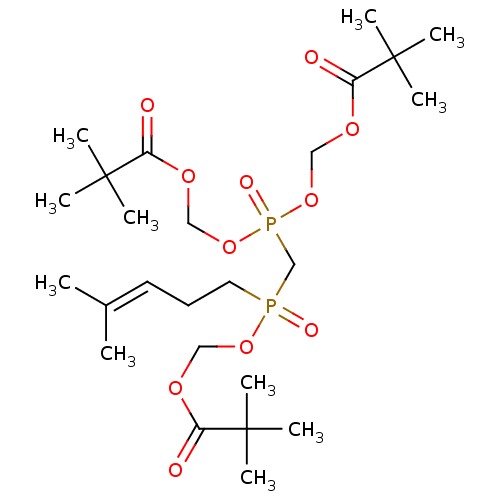
BDBM50236891 CHEMBL4081059
SMILES [#6]\[#6](-[#6])=[#6]\[#6]-[#6]P(=O)([#6]P(=O)([#8]-[#6]-[#8]-[#6](=O)C([#6])([#6])[#6])[#8]-[#6]-[#8]-[#6](=O)C([#6])([#6])[#6])[#8]-[#6]-[#8]-[#6](=O)C([#6])([#6])[#6]
InChI Key InChIKey=DUNPBMOQWISNAG-UHFFFAOYSA-N
Data 2 EC50
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50236891
Found 2 hits for monomerid = 50236891
Affinity DataEC50: >1.00E+5nMAssay Description:Activation of BTN3A1 in human Vgamma9Vdelta2 T cells assessed as induction of interleukin 2 stimulated cell proliferation treated for 72 hrs measured...More data for this Ligand-Target Pair
Affinity DataEC50: 7.30E+3nMAssay Description:Activation of BTN3A1 in human Vgamma9Vdelta2 T cells assessed as induction of T effector cells mediated K562 cell lysis preincubated with K562 cells ...More data for this Ligand-Target Pair