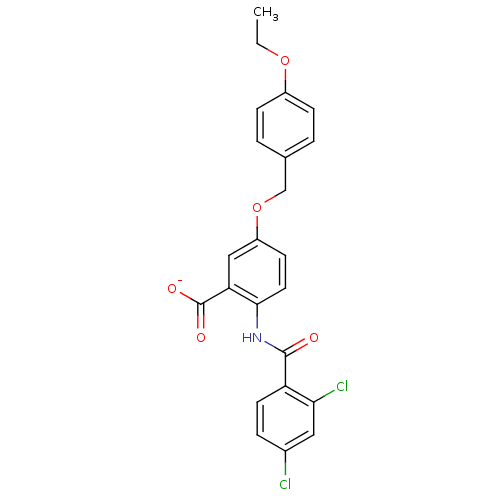
BDBM50121419 CHEMBL119798::Lithium; 2-(2,4-dichloro-benzoylamino)-5-(4-ethoxy-benzyloxy)-benzoate
SMILES CCOc1ccc(COc2ccc(NC(=O)c3ccc(Cl)cc3Cl)c(c2)C([O-])=O)cc1
InChI Key InChIKey=NLLLBTAOCGRTOK-UHFFFAOYSA-M
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50121419
Found 4 hits for monomerid = 50121419
Affinity DataEC50: >1.00E+5nMAssay Description:Transactivation potency was measured by luciferase activity in Caco- 2/TC7 cells transiently co-transfected for the fusion-protein Gal4-PPAR alphaMore data for this Ligand-Target Pair
Affinity DataEC50: 700nMAssay Description:Transactivation potency was measured by luciferase activity in Caco- 2/TC7 cells transiently co-transfected for the fusion-protein Gal4-PPAR gammaMore data for this Ligand-Target Pair
Affinity DataKi: 3.60E+3nMAssay Description:Ligand binding affinity was determined by displacement of a tritiated tracer by the unlabeled compound to a GST-PPAR fusion protein for Peroxisome pr...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Transactivation potency was measured by luciferase activity in Caco- 2/TC7 cells transiently co-transfected for the fusion-protein Gal4-PPAR alphaMore data for this Ligand-Target Pair