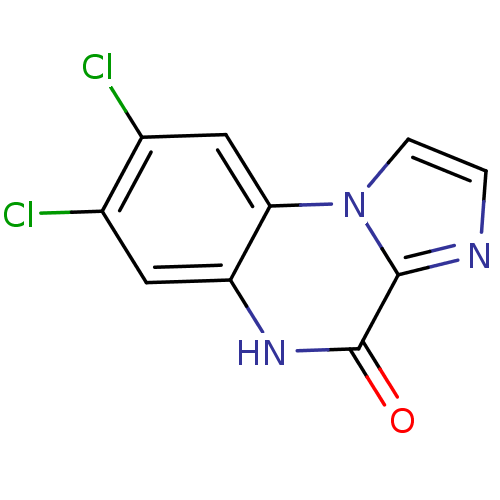
BDBM50002696 7,8-Dichloro-5H-imidazo[1,2-a]quinoxalin-4-one::CHEMBL325124
SMILES Clc1cc2[nH]c(=O)c3nccn3c2cc1Cl
InChI Key InChIKey=INRPWJDBSCEMAX-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50002696
Found 4 hits for monomerid = 50002696
Affinity DataIC50: 1.00E+5nMAssay Description:Compound was evaluated for its binding affinity towards strychnine - insensitive glycine site of NMDA in presence of [3H]- CPPMore data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+3nMAssay Description:Compound was evaluated for its binding affinity towards strychnine - insensitive glycine site of NMDA receptor in presence of [3H]- GlyMore data for this Ligand-Target Pair
Affinity DataIC50: 1.26E+3nMAssay Description:Displacement of [3H]glycine from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 1.26E+3nMAssay Description:Displacement of [3H]glycine from glycine site on the NMDA receptor.More data for this Ligand-Target Pair