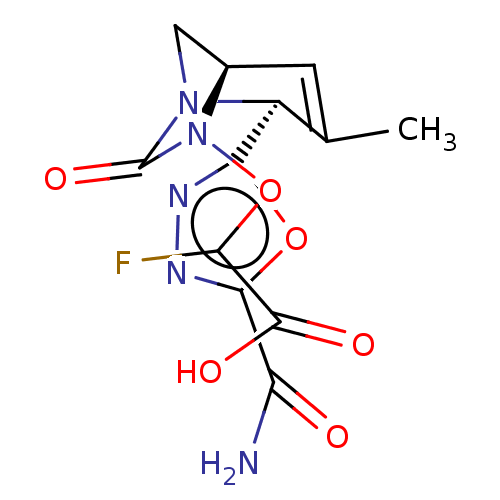
BDBM466977 2-[[(2S,5R)-2-(5-carbamoyl-1,3,4-oxadiazol-2-yl)-3-methyl-7-oxo-1,6-diazabicyclo[3.2.1]oct-3-en-6-yl]oxy]-2-fluoro-acetic acid lithium salt::US10800778, Example 55
SMILES CC1=C[C@@H]2CN([C@@H]1c1nnc(o1)C(N)=O)C(=O)N2OC(F)C(O)=O
InChI Key InChIKey=YHANUKIZKPOWPW-UHFFFAOYSA-N
Data 3 IC50
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 466977
Found 3 hits for monomerid = 466977
Affinity DataIC50: 7.30nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
Affinity DataIC50: 24nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair
Affinity DataIC50: 180nMAssay Description:A buffer consisting of 0.1 M sodium phosphate (pH 7.0), 10 mM NaHCO3, and 0.005% Triton X-100 was used for all enzymes. The chromogenic substrate nit...More data for this Ligand-Target Pair