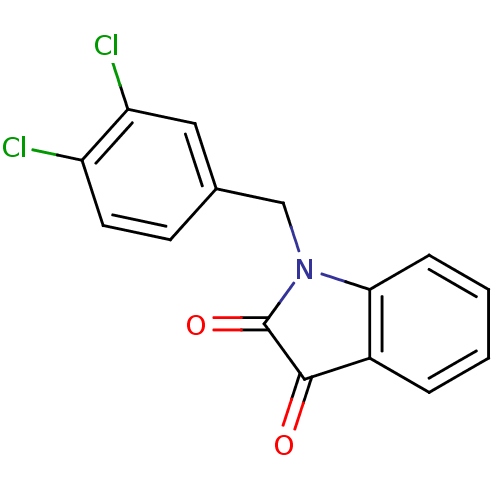
BDBM22796 1-[(3,4-dichlorophenyl)methyl]-2,3-dihydro-1H-indole-2,3-dione::Isatin-based compound, 16::cid_1901244
SMILES Clc1ccc(CN2C(=O)C(=O)c3ccccc23)cc1Cl
InChI Key InChIKey=KGRJPLRFGLMQMV-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 7 hits for monomerid = 22796
Found 7 hits for monomerid = 22796
Affinity DataKi: 31nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 65nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 67nM ΔG°: -9.78kcal/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.26E+4nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataIC50: 2.27E+4nMAssay Description:Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBIMR, San Diego, C...More data for this Ligand-Target Pair
TargetApoptotic protease-activating factor 1(Human)
Burnham Center For Chemical Genomics
Curated by PubChem BioAssay
Burnham Center For Chemical Genomics
Curated by PubChem BioAssay
Affinity DataIC50: 3.40E+4nMAssay Description:Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBIMR, San Diego, C...More data for this Ligand-Target Pair
Affinity DataKi: 4.82E+4nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair