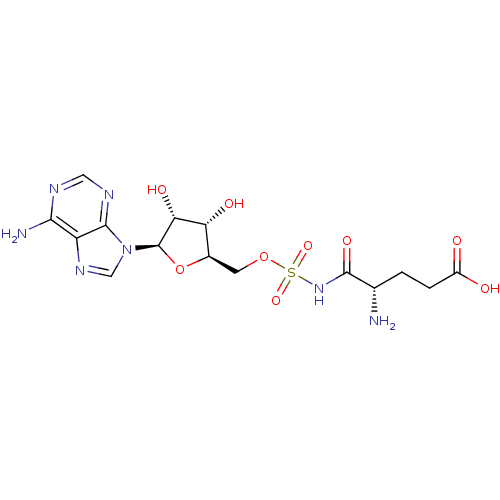
BDBM18122 (4S)-4-amino-5-[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}sulfonyl)amino]-5-oxopentanoic acid::5-O-[N-(L-Glutamyl)-sulfamoyl]adenosine 5::Glu-AMS
SMILES N[C@@H](CCC(O)=O)C(=O)NS(=O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(N)ncnc12
InChI Key InChIKey=YBRKRYFZKHICLS-UHFFFAOYSA-N
Data 2 KI
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 18122
Found 2 hits for monomerid = 18122
Affinity DataKi: 2.80nM ΔG°: -12.1kcal/molepH: 7.2 T: 2°CAssay Description:The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p...More data for this Ligand-Target Pair
Affinity DataKi: 70nM ΔG°: -9.92kcal/molepH: 8.0 T: 2°CAssay Description:The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p...More data for this Ligand-Target Pair