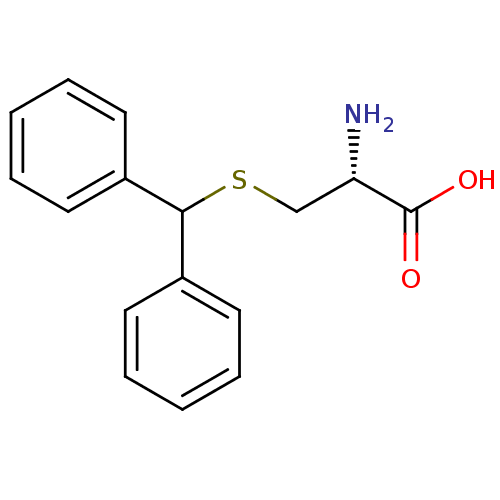
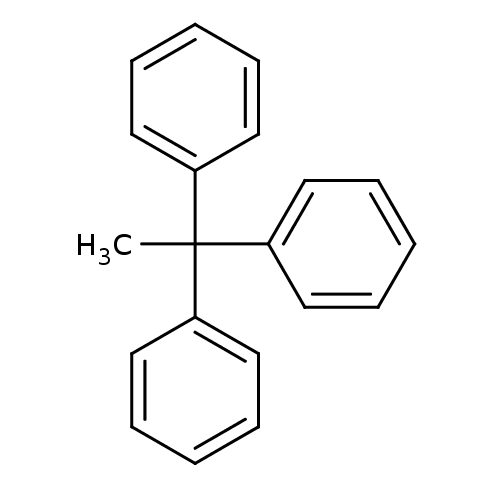
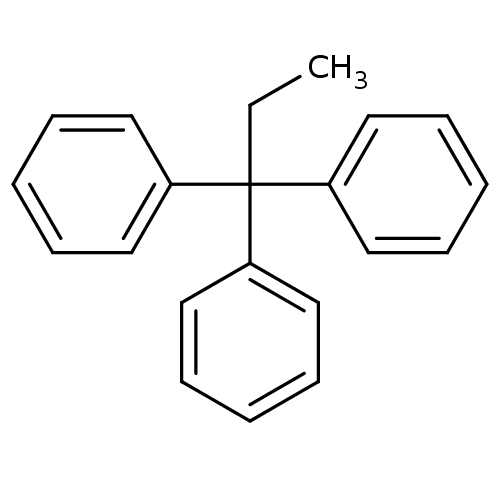
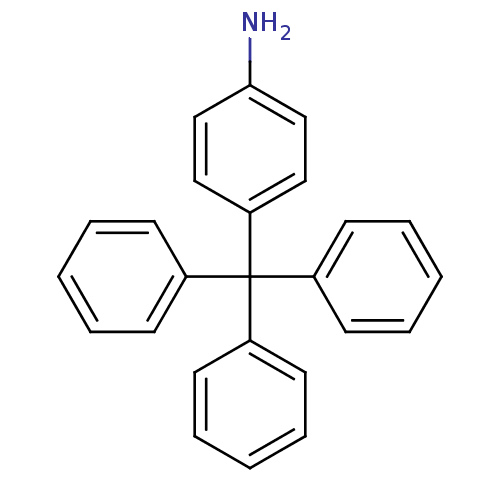
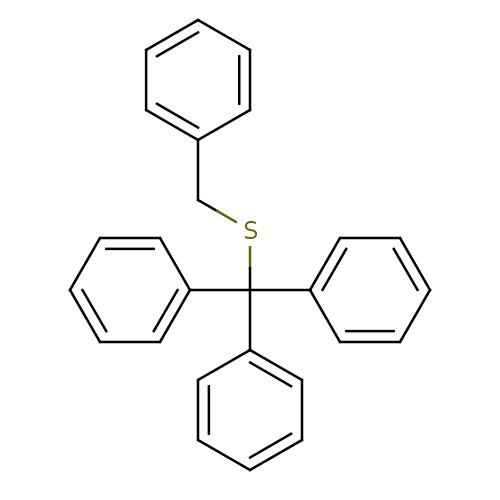
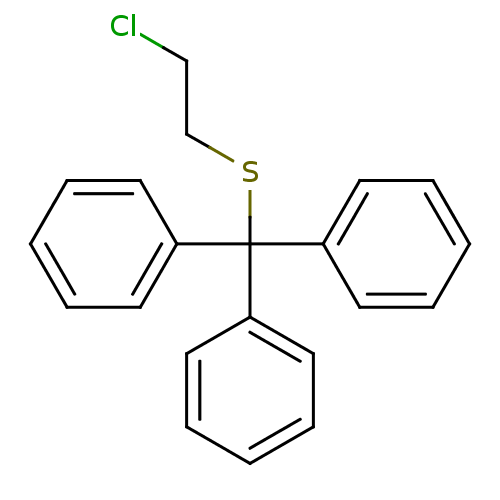
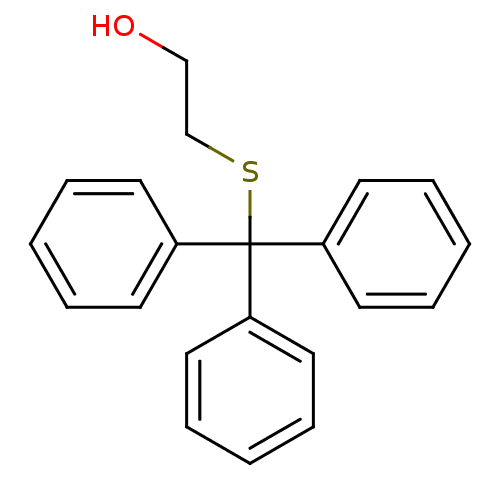
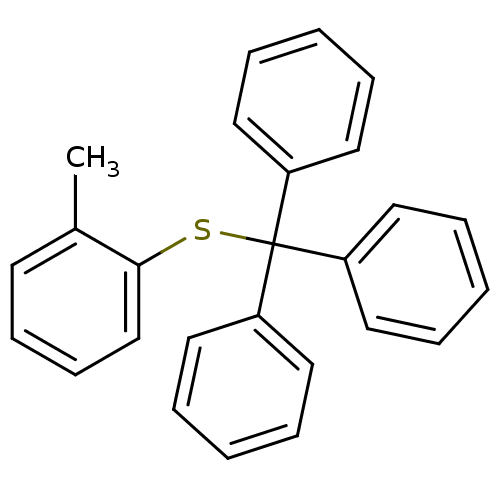
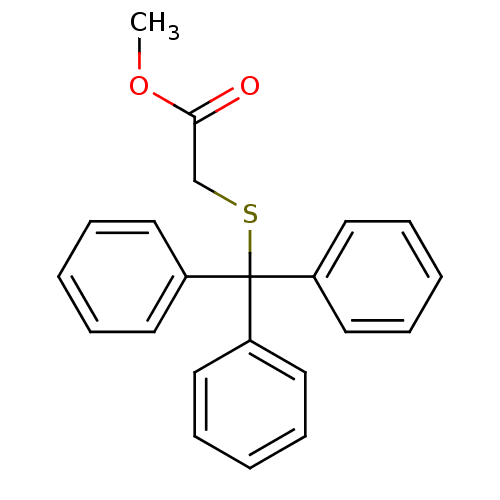
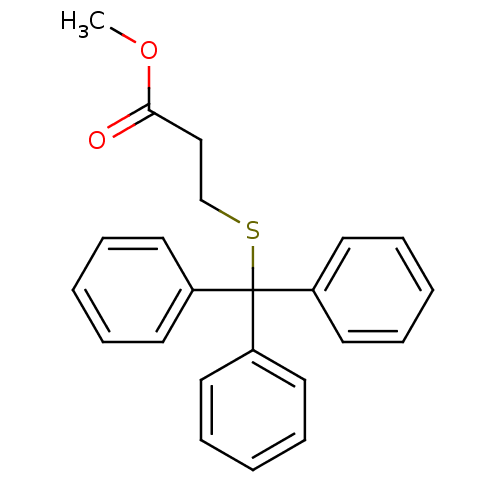
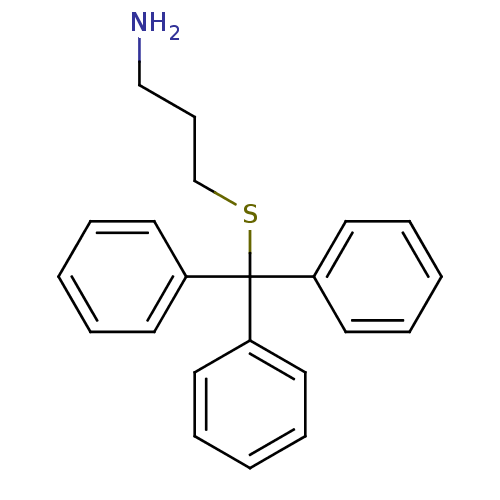
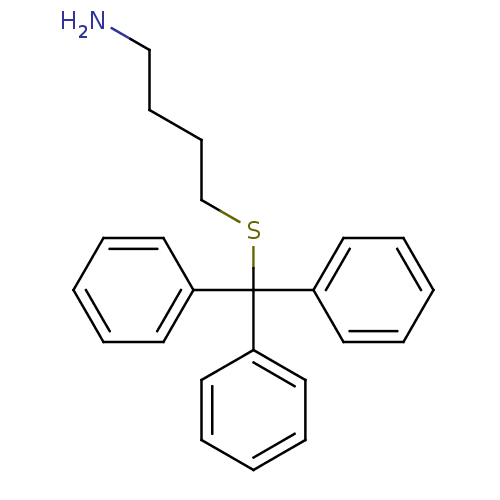
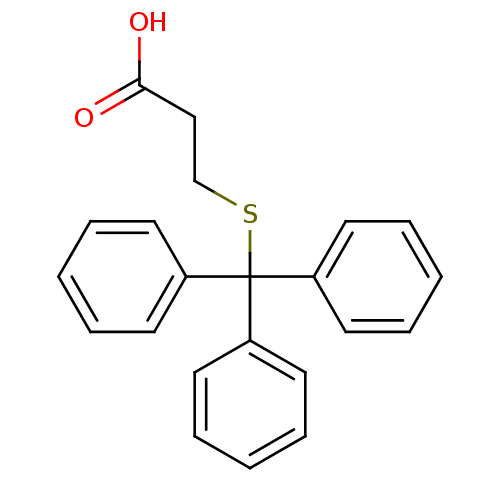
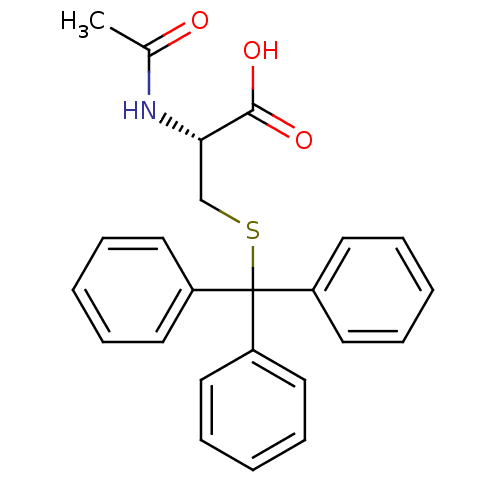
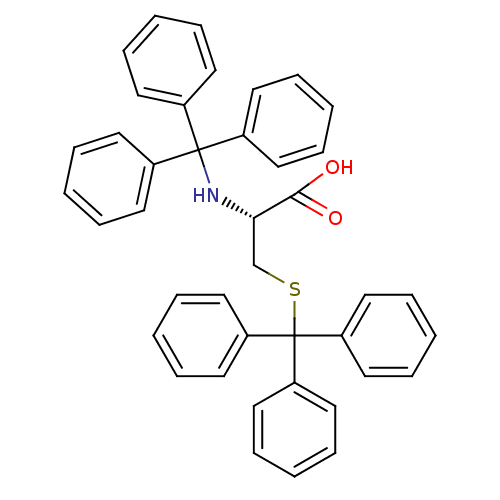
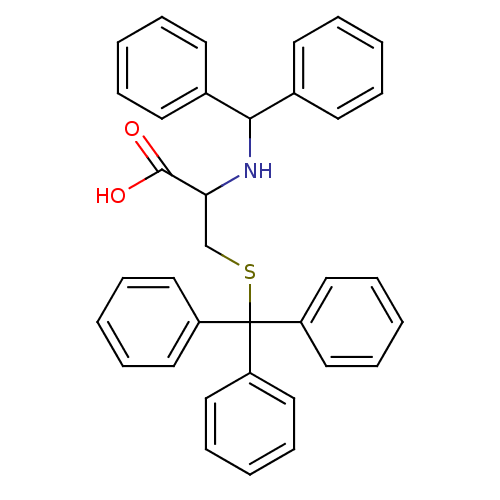
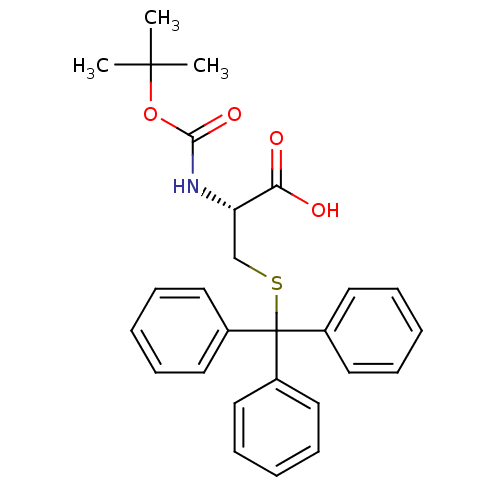
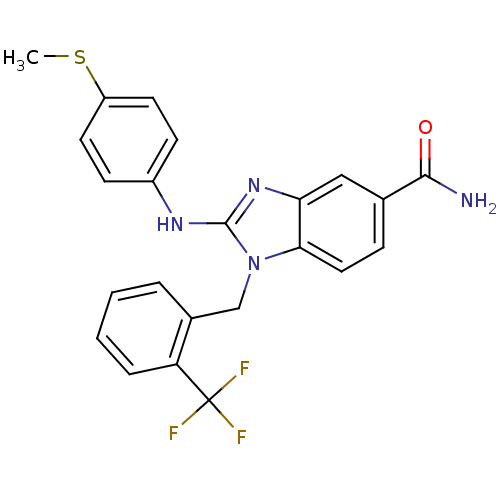
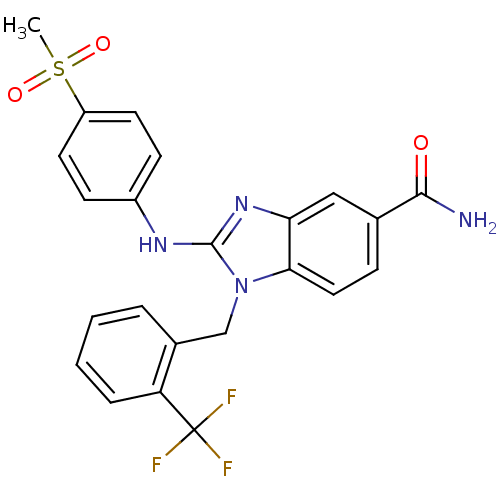
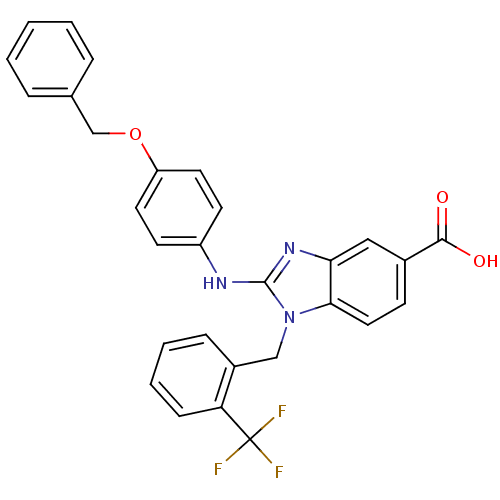
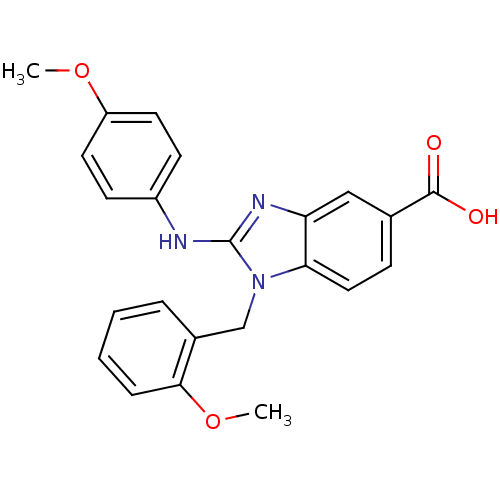
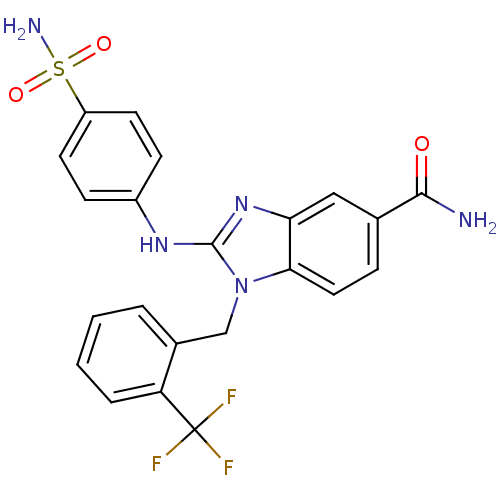
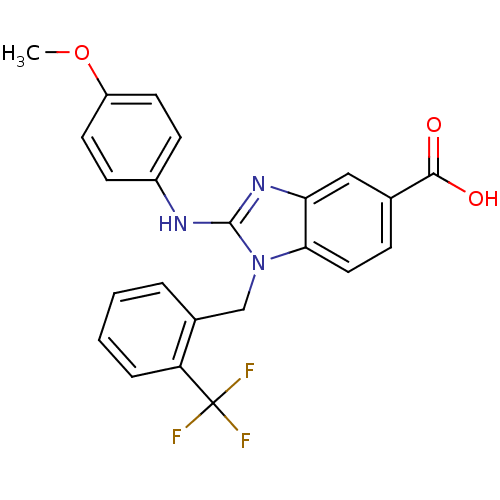
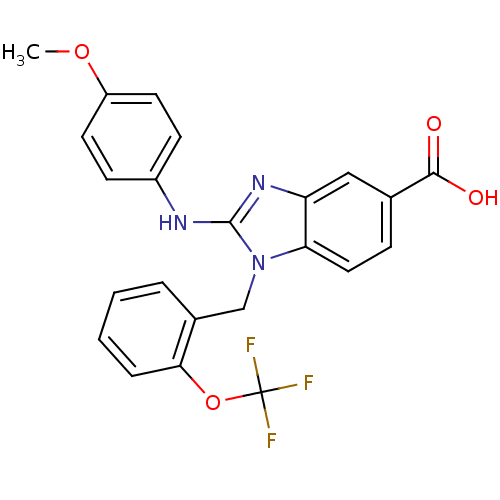
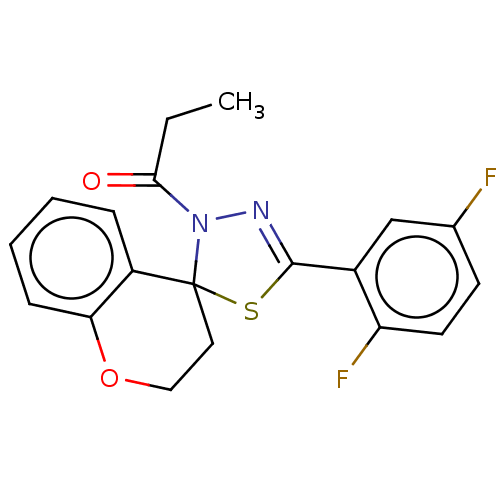
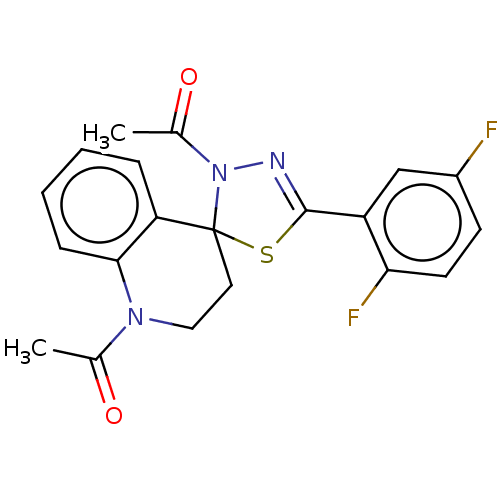
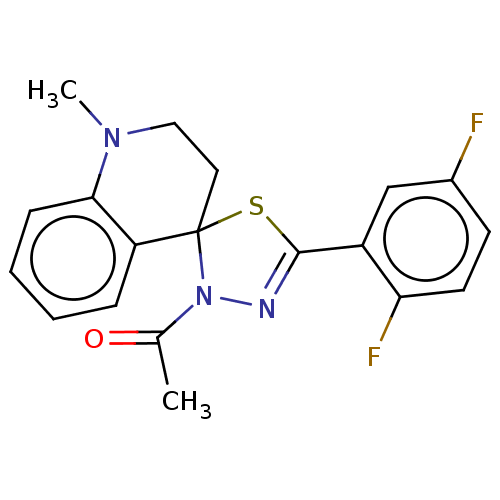
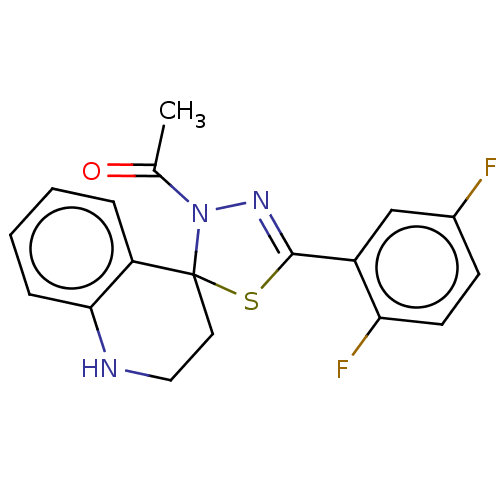
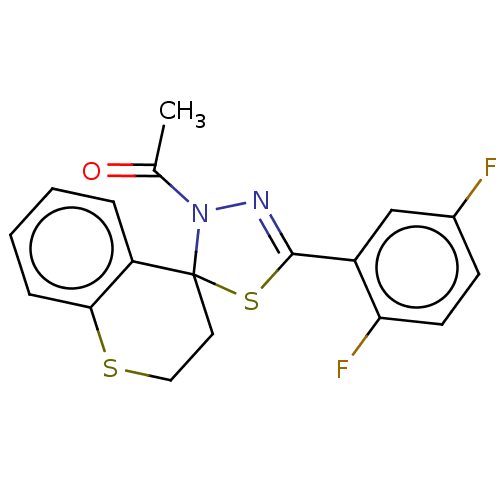
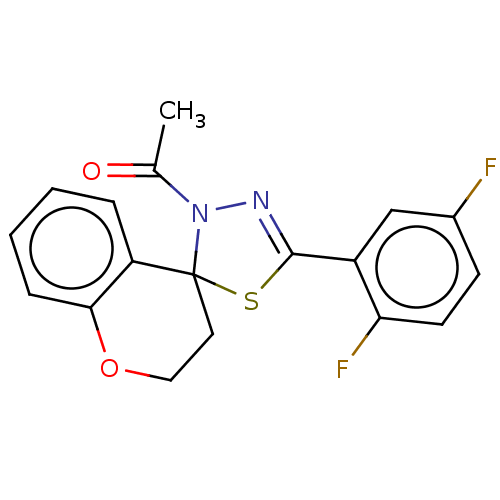
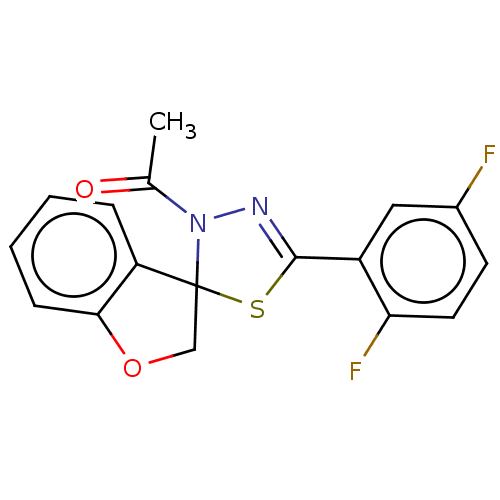
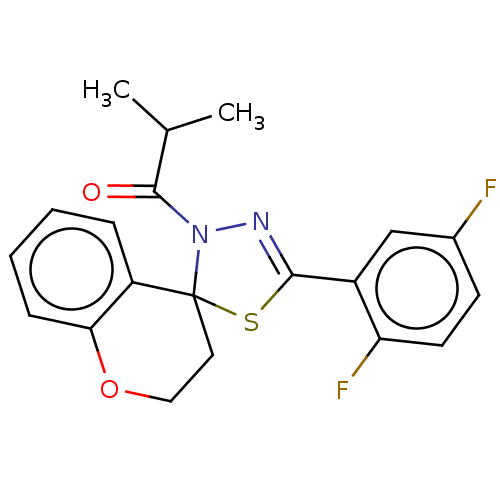
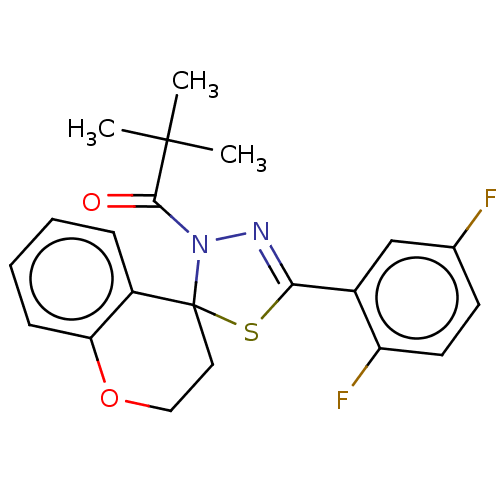
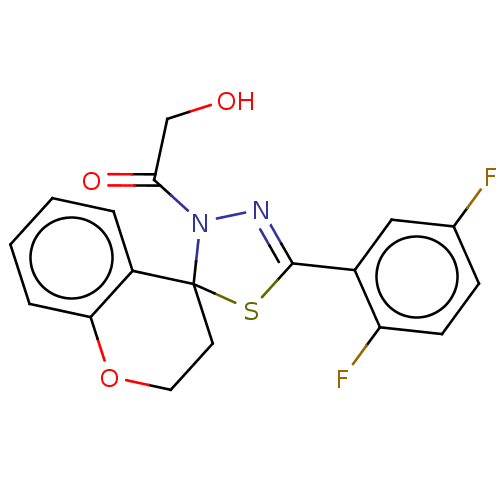
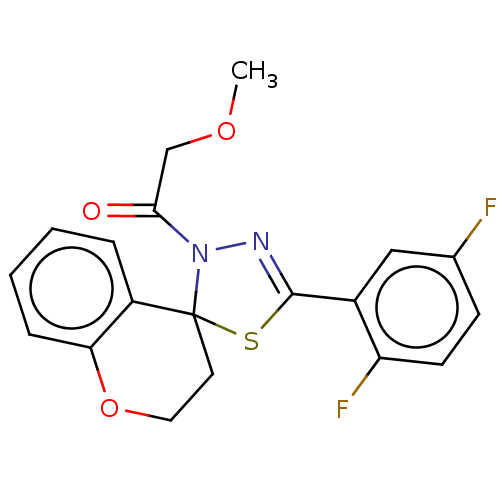
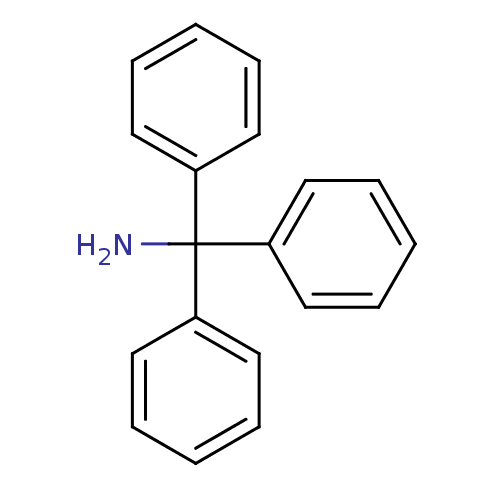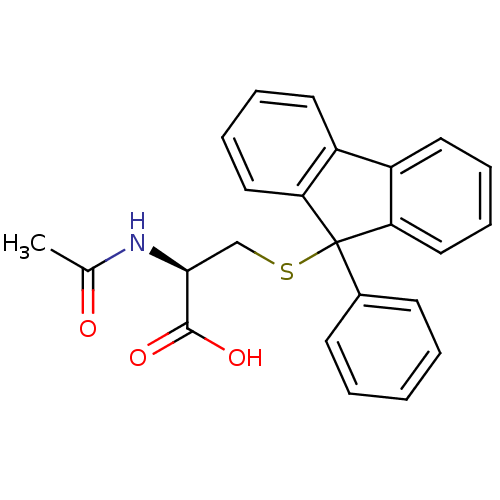Affinity DataKd: >10nMAssay Description:Binding affinity to human wild type Eg5 at 250 uM by isothermal calorimetric analysisMore data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataT: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair
Affinity DataKd: 2.00E+3nMAssay Description:Binding assay using temperature-dependent circular dichroism (TdCD). In TdCD, the loss of protein secondary structure was monitored as a function of...More data for this Ligand-Target Pair
Affinity DataKd: 2.50E+3nMAssay Description:Binding assay using temperature-dependent circular dichroism (TdCD). In TdCD, the loss of protein secondary structure was monitored as a function of...More data for this Ligand-Target Pair
Affinity DataKd: 1.40E+4nMAssay Description:Binding assay using temperature-dependent circular dichroism (TdCD). In TdCD, the loss of protein secondary structure was monitored as a function of...More data for this Ligand-Target Pair
Affinity DataKd: 5.00E+3nMAssay Description:Binding assay using temperature-dependent circular dichroism (TdCD). In TdCD, the loss of protein secondary structure was monitored as a function of...More data for this Ligand-Target Pair
Affinity DataKd: 600nMAssay Description:Binding assay using temperature-dependent circular dichroism (TdCD). In TdCD, the loss of protein secondary structure was monitored as a function of...More data for this Ligand-Target Pair
Affinity DataKd: 1.20E+3nMAssay Description:Binding assay using temperature-dependent circular dichroism (TdCD). In TdCD, the loss of protein secondary structure was monitored as a function of...More data for this Ligand-Target Pair
Affinity DataKd: 3.40E+3nMAssay Description:Binding assay using temperature-dependent circular dichroism (TdCD). In TdCD, the loss of protein secondary structure was monitored as a function of...More data for this Ligand-Target Pair
Affinity DataEC50: 452nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+3nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+3nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: 179nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: 3.31E+3nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+3nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: 139nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: 246nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: >500nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+3nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: 615nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
Affinity DataEC50: 665nMAssay Description:Inhibition of KSP in human HCT116 cells assessed as phos-histone H3 level by immunofluorescent assayMore data for this Ligand-Target Pair
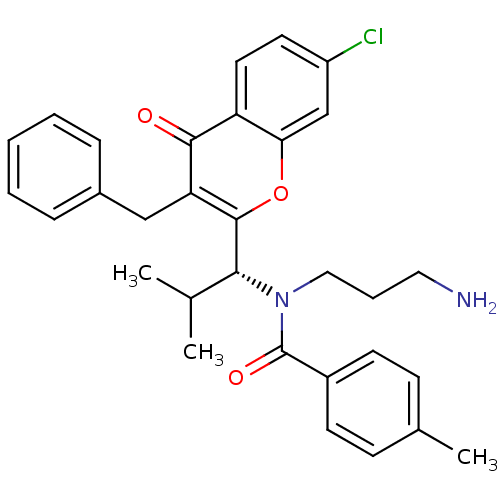
 3D Structure (crystal)
3D Structure (crystal)