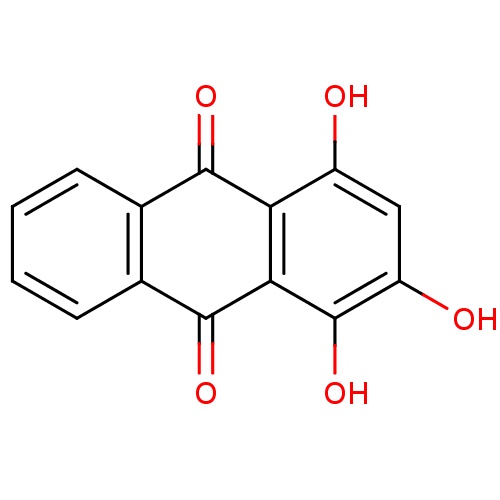
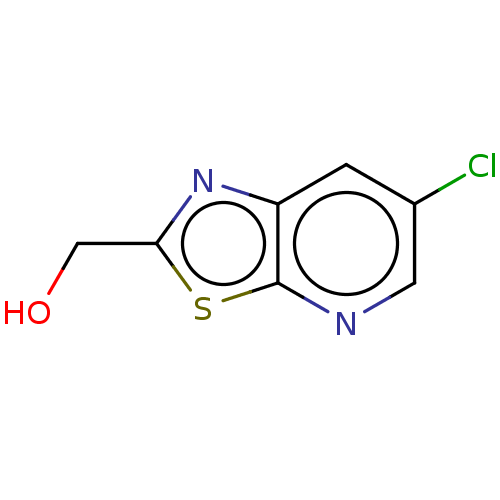
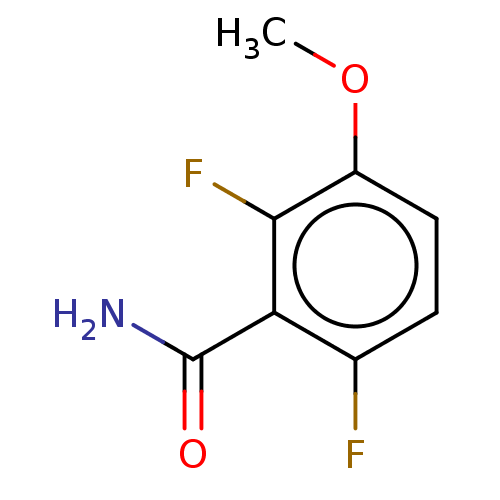
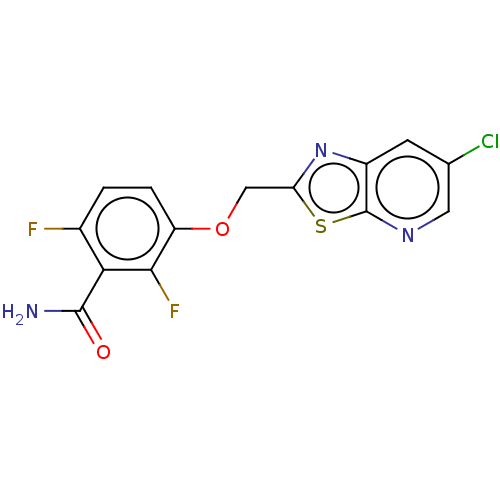
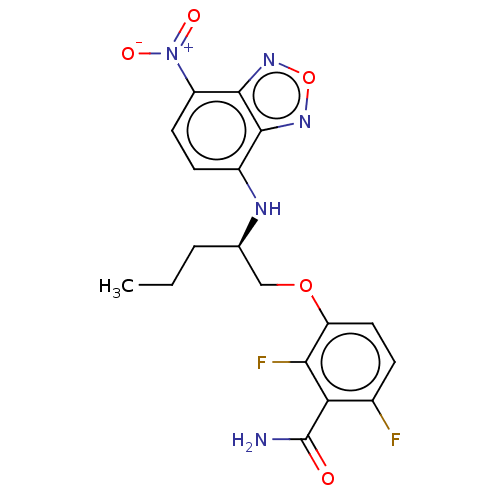
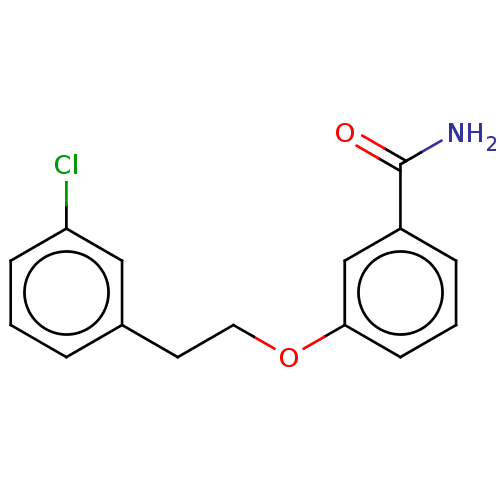
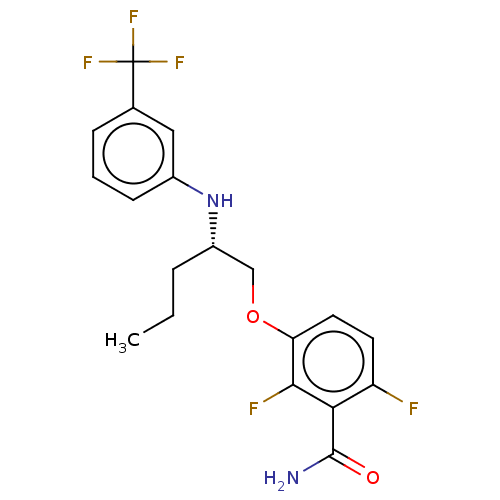
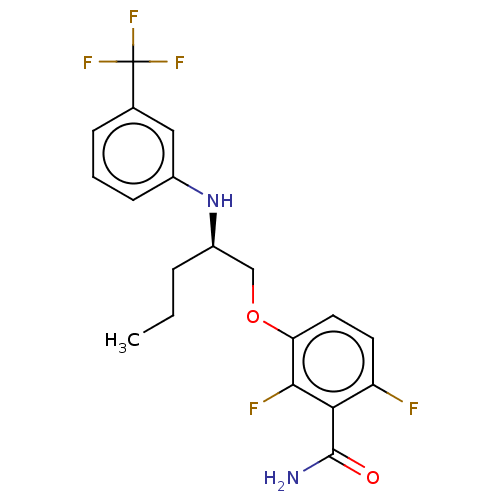
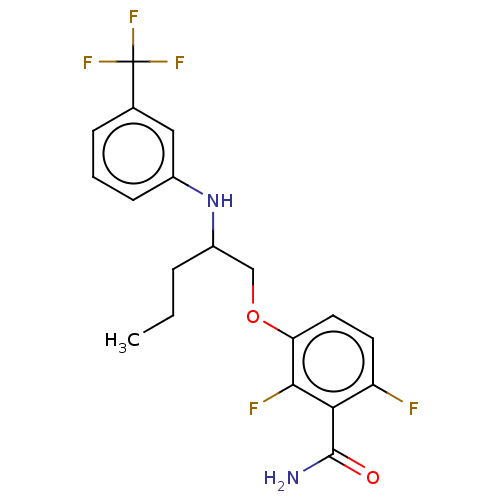
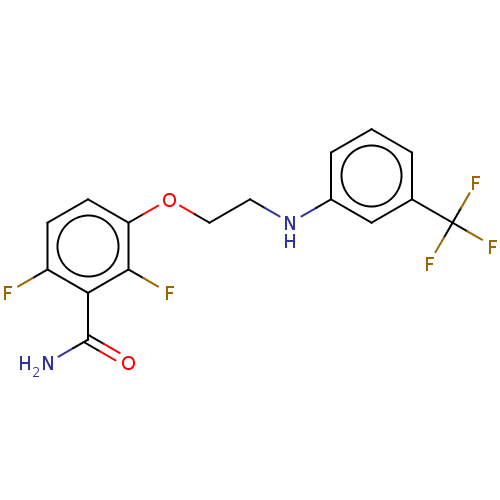
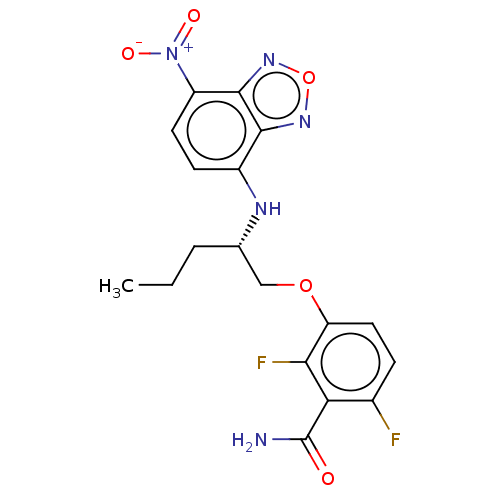
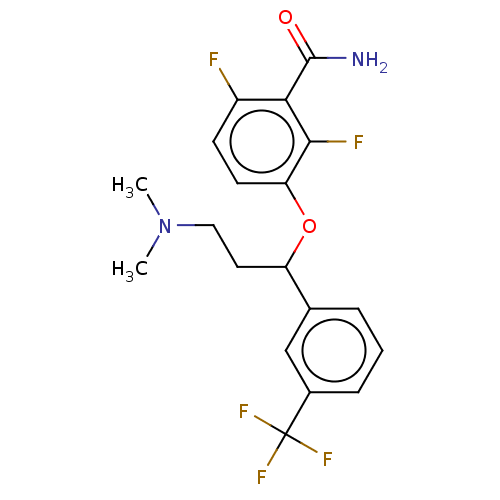
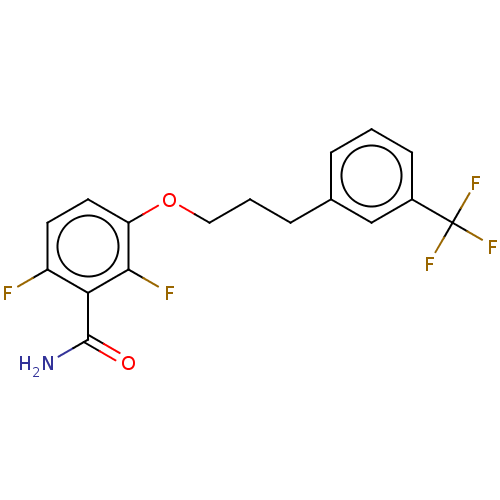
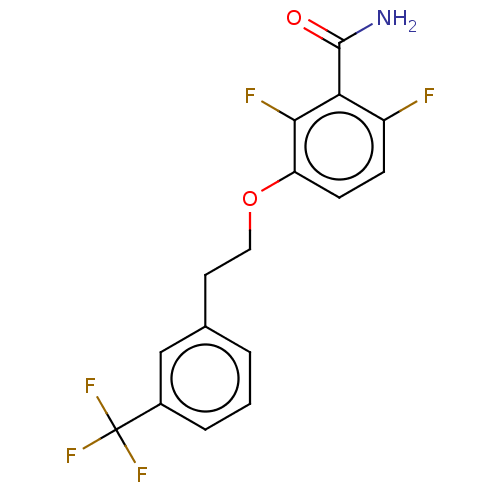
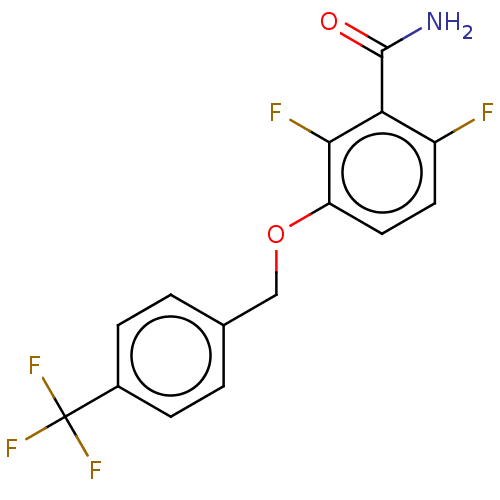
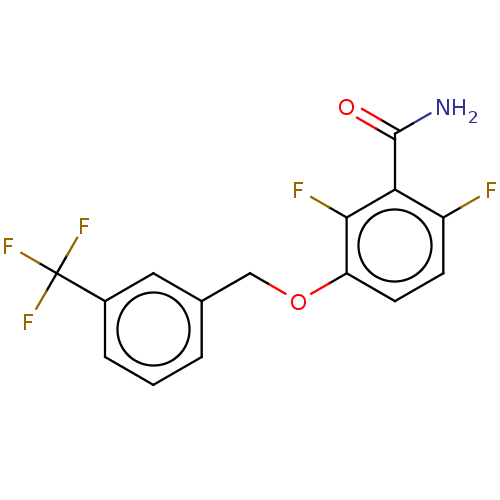
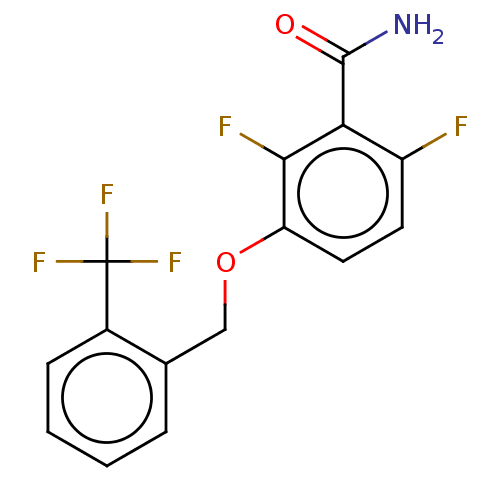
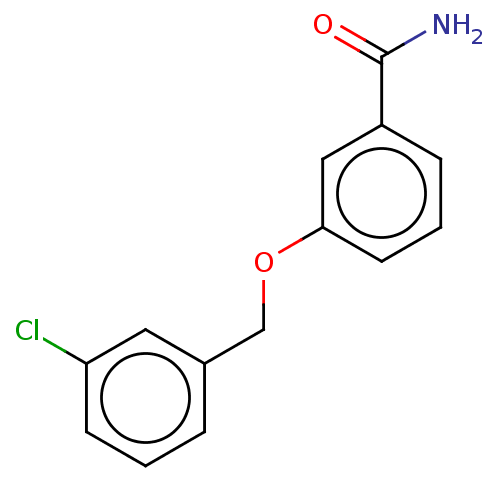
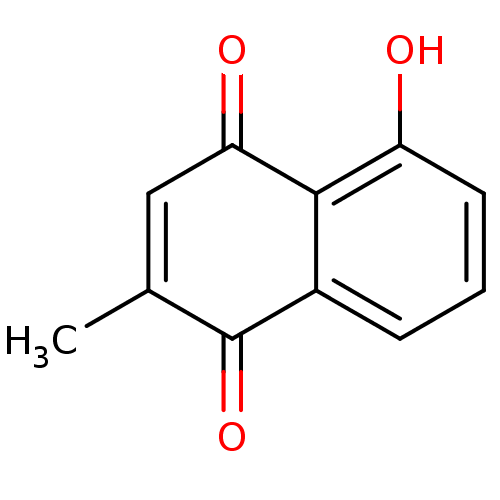
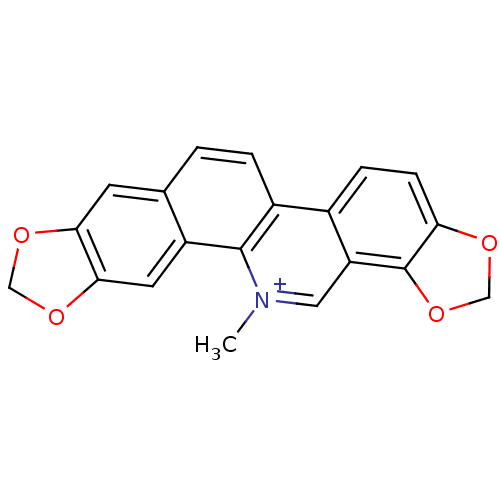
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 5.20E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ assessed as dissociation constant of intrinsic tryptophan quenching incubated for 30 mins by double recipr...More data for this Ligand-Target Pair
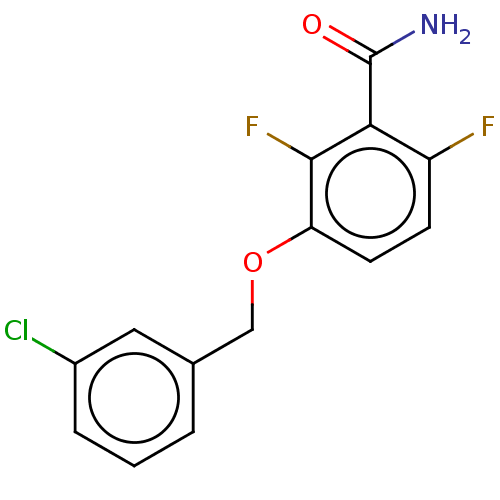
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.00E+6nMAssay Description:Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ...More data for this Ligand-Target Pair
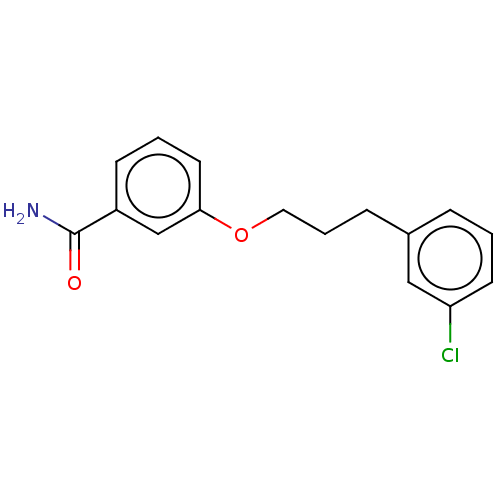
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 6.70E+5nMAssay Description:Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ...More data for this Ligand-Target Pair
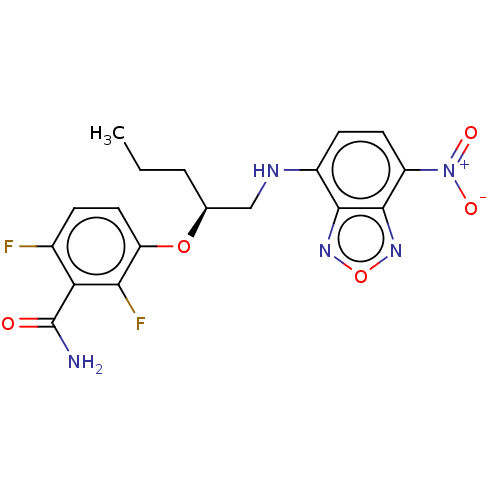
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: <100nMAssay Description:Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ...More data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.50E+4nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 7.10E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 3.80E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 200nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.10E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.45E+5nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 5.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 6.70E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 700nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 7.10E+4nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.30E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 9.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 8.00E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 9.30E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataKd: 1.58E+4nMAssay Description:Binding affinity to Bacillus subtilis FtsZ assessed as dissociation constant of intrinsic tryptophan quenching incubated for 30 mins by double recipr...More data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataIC50: 3.00E+3nMAssay Description:Inhibition of FtsZ in Bacillus subtilis assessed as disruption of Z ring formationMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis)
National Institute of Technology Calicut
Curated by ChEMBL
National Institute of Technology Calicut
Curated by ChEMBL
Affinity DataIC50: 7.00E+4nMAssay Description:Inhibition of Bacillus subtilis FtsZ GTPase activity assessed as production of inorganic phosphate measured up to 60 mins by malachite green-phosphom...More data for this Ligand-Target Pair